Introduction
Plasmodium falciparum parasites evidence genetic diversity in relation to their geographical origin. Several P.falciparum genes show extensive genetic polymorphisms and the diversity spectrum has been more fully characterized in parasites from the African continent. Plasmodium falciparum represents the second most frequent species after P. vivax in the Americas. The highest rates of infection are reported in regions inhabited primarily by persons of African descent. In Colombia, these settlements are along the Pacific coast and in Northwestern Colombia (Uraba, Bajo Cauca and Cordoba regions).
The genetic characteristics of particular polymorphic genes, such as msp1, msp2 and glurp 1 - 3 , have been applied to explore parasite population dynamics in malaria endemic areas. In areas of intense transmission, msp1, msp1 and glurp have proved very polymorphic and highly effective in discriminating recrudescent from re-infecting alleles 4 . In the endemic regions of America, variability using these markers has reported uneven results 5 . However, in Colombia, the genes appear disseminated across the country with only minor differences in polymorphism frequencies 6 - 8 . Major climate changes have affected our country during the past 10 years. This has been seen mainly by extremes in the dry and rainy seasons that have resulted from the El Niño/Southern Oscillation (ENSO) 9 . This can have severe consequences on the prevalence and dissemination of particular genotypes. For example, resistant clones might easily spread through regions that share the same eco-epidemiological area after there have been major changes in mosquito populations and intensity rates.
In addition, the implementation in 2006 of a nationwide policy of artemisinin-combined therapy against falciparummalaria resulted in a new challenge for health authorities: to provide rapid diagnoses and effective treatments in a country where political instability is widespread in the malaria endemic regions.
Treatment with artemisinin-based schemes further affects the genotype of P. falciparum populations by introducing new drug pressures on parasites already resistant to other anti-malarials 10 . In this context, exploration of the genetic characteristics of this species has been a major focus of our research.
In order to establish the baseline genetic composition of P. falciparum, clinical isolates were identified before the introduction of artemisinin-combined treatments from the regions with the highest frequency of malarial infection in the country. In order to provide adequate genetic information for future studies that will explore changes in population structures, we are reporting the genotypes of all archived samples collected from 2004 to 2008 in Northwest Colombia. This was done as part of a nationwide study aimed at monitoring the presence of anti-malarial resistance.
Materials and Methods
Study sites and sample design
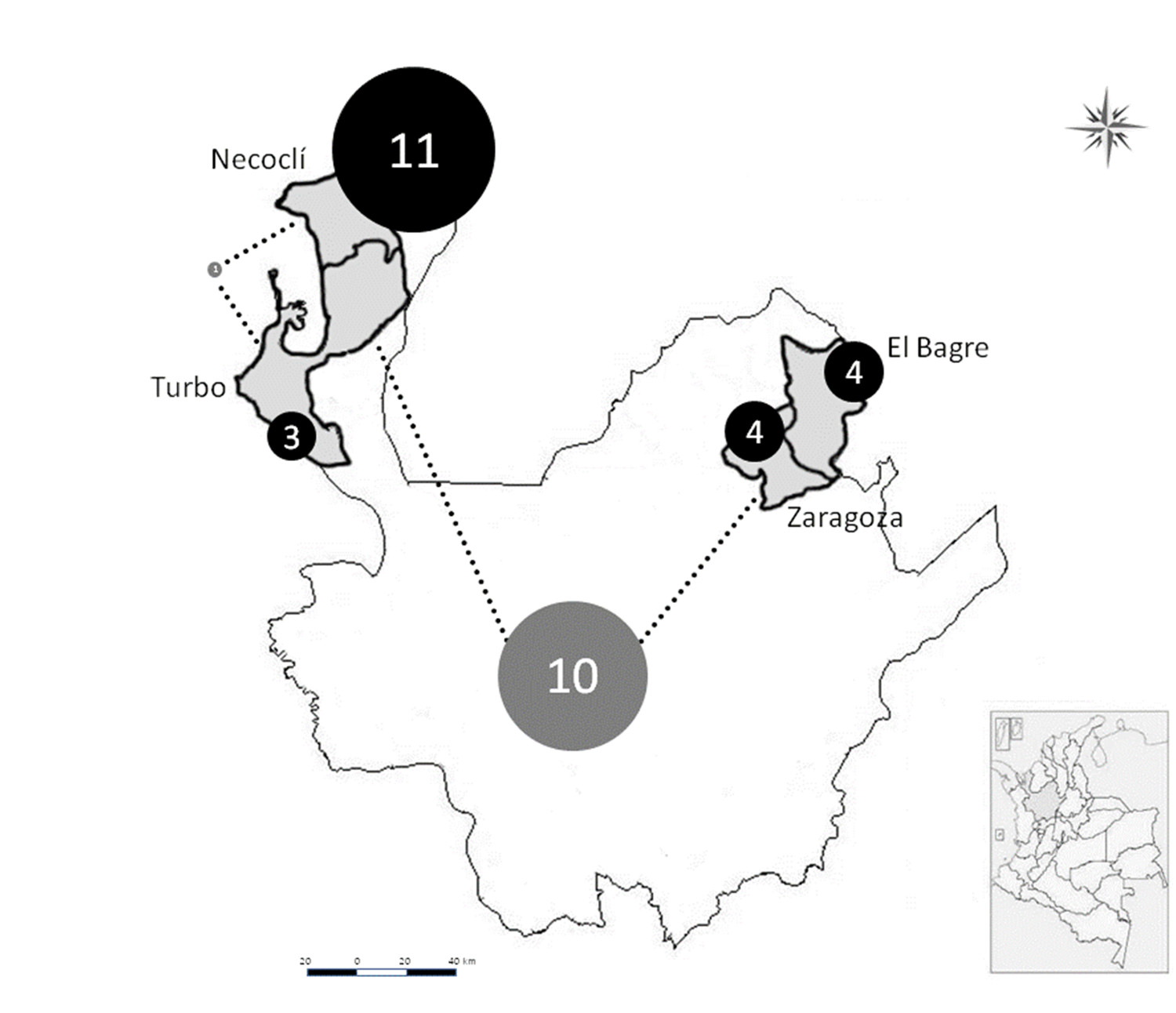
This study was part of a major anti-malarial efficacy trial that took place in the municipalities of Necocli (8º,25´ 11´´ N, 76º 45´ 58´´ W), Turbo (8° 5´4´´ N, 76° 44´ 123´´ W), El Bagre (7° 35´ 2´´ N, 74° 48´ 2´´ W) and Zaragoza (7° 29´ 2´´ N, 74° 51´ 35´´ W). These are located in two regions of Northwestern Colombia where malaria is highly endemic: Urabá and Bajo Cauca. Both regions have: a) high migration rates and perennial malarial transmission that was unstable with mean annual parasite index (API: numbers of cases per 1,000 inhabitants) - during 2004-2008 of 61.47 in Necocli, 33.09 in Turbo, 128.48 in El Bagre and 127.04 in Zaragoza 11 - 13 ; b) these are mainly inhabited by African descendants and persons with indigenous ancestry, as well as by those with mixed indigenous and Spanish ancestry; c) the economy of Bajo Cauca is based on gold mining, whereas Urabá is a banana-producing region 14 , 15 .
The macro-project was designed as a random, clinically-controlled study in which anti-malarial efficacy was assessed on the basis of the mean incidence of falciparum malaria for each 10,000 cases per year (50% in each region) with an estimated 20% maximum treatment failure rate, 95% confidence interval, 5% sampling error. The current report presents the results for all subjects recruited in the original study from which genetic material was available.
The study included patients attending the local malaria clinics with an acutely symptomatic, non-severe form of P. falciparum malaria. Unique infection by this species was confirmed by microscopy and by nested PCR. The inclusion criteria for the original study were: ≥1 year of age, unique P. falciparum parasitaemia of ≥1,000 asexual forms/µL, and the willingness to participate. Patients were excluded if consent was withdrawn.
The study protocol was reviewed and approved by the Ethics Committee of the Faculty of Medicine, Universidad de Antioquia (Medellín, Colombia). Each participant gave their fully informed consent.
Diagnosis of P. falciparum infection
Whole blood was taken from a peripheral vein on day 0 before the administration of treatment. Giemsa-Field stained thick/thin blood films were examined at 100x magnification to identify the presence of parasites, Plasmodium species, parasite density, and schizontaemia.
Parasite density was measured by counting the number of asexual parasites per 200 leukocytes based on a mean count of 8,000 leukocytes per microliter of blood (theoretical value) 16 . A slide was considered negative after the examination of at least 300 microscopic fields 16 . In addition, a nested PCR was performed to confirm unique infection by P. falciparum. Amplification from whole blood, collected and extracted (as detailed in the Molecular genotyping section), was carried out using a nested PCR assay to detect the 18s ribosomal RNA (rRNA) gene of P. falciparum and P. vivax, according to previously published procedures 17 . Amplification products were resolved in a 2% agarose gel using ethidium bromide and viewing under UV light.
Molecular genotyping
Only pre-treatment samples (day 0) from sequentially recruited subjects were selected for the genotyping of msp1, msp2, and glurp.
A volume of 100 mL of whole blood was placed on Whatman 3MM filter paper and stored at -20○ C until DNA extraction with Chelex(r). Briefly, a piece of filter paper was soaked overnight in a solution of 10% saponin in PBS and was subsequently washed in PBS. Afterwards, genomic DNA from each sample was obtained by boiling, followed by ethanol precipitation that was conducted according to established procedures 18 .
The region 2 of msp1 and msp2 (central region) were amplified by a nested PCR; glurp (region R2) was amplified by a semi-nested PCR 19 . Products obtained after the first PCR, were amplified using specific primers for region 2 of msp1 corresponding to MAD20, K1, and RO33 allellic families, and FC27 and IC-1 for the central region of msp2. The sequence of the primers and the protocol of PCR are described in detail by Snounou et al 19 . Briefly, a PCR was carried out in a total volume of 20 µL, containing 10 mM Tris-HCl (pH 9.0 at 25○ C), 50 mMKCl 134 and 0.1% Triton X-100, 125 mMdNTPs, 0.4 units Taq DNA polymerase (Fermentas), 1.6 mM MgCl2, and 125 nM of each primer. Initial denaturation was for 5 min at 95○ C, 1 min at 94○ C, 2 min at 58○ C annealing (all first PCRs and secondary reactions for glurp) or at 61○ C (for all the secondary reactions of msp1 and msp2). This was followed by an extension for 2 min at 72○ C. This first reaction underwent 25 amplification cycles and the secondary 30 cycles.
Positive (strains HB3, K1, and RO33) and negative controls (healthy individuals) were included. Products were electrophoresed on agarose (MetaPhor) gel (2.5% for msp1 and 2% for msp2 and glurp), stained with ethidium bromide, and read under ultraviolet light. Size polymorphism for each gene was assessed using the Quantity One (Bio-Rad) program.
Results
In total, genetic material was available from 224 subjects from the two regions: 136 in Uraba (Turbo n= 95, Necocli n= 41) and 88 in Bajo Cauca, (El Bagre n= 75, Zaragoza n= 13). From this sample, all (100%) were successfully amplified for msp1, 205 (91%) for msp2 and 172 (77%) for glurp.
Genetic diversity and allelic frequency
For msp1, five different genotypes were observed: representing the K1 (2 genotypes), MAD20 (1 genotype) and RO33 (2 genotypes) allelic families. The msp1 fragment sizes ranged from 150-270 bp. However, all samples corresponded to the monomorphic MAD20 150 bp genotype (Table 1) while the K1 (150-200 bp) and RO33 (250-270 bp) allelic families were detected below the level of 5%.
Table 1
Comparison of genotypes according to the msp1, mps2, and glurp genes in the four localities examined. Results are compared according to the year of sample collection as reported by different authors in the periods of 2000-2004, 2004-2006 20-23, and 2006-2008 (the current study). Alleles corresponding to the FC27 family of msp2 were absent in all studies
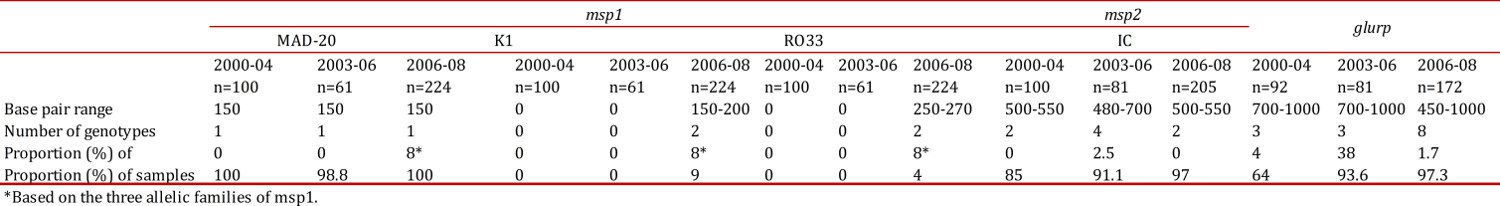
A total of 2 different msp2 genotypes (size 500 and 550 pb) were recorded in 91% of the samples and they were all from the IC allelic family. In this marker, an 85% frequency rate was detected for the fragment of 500 bp.
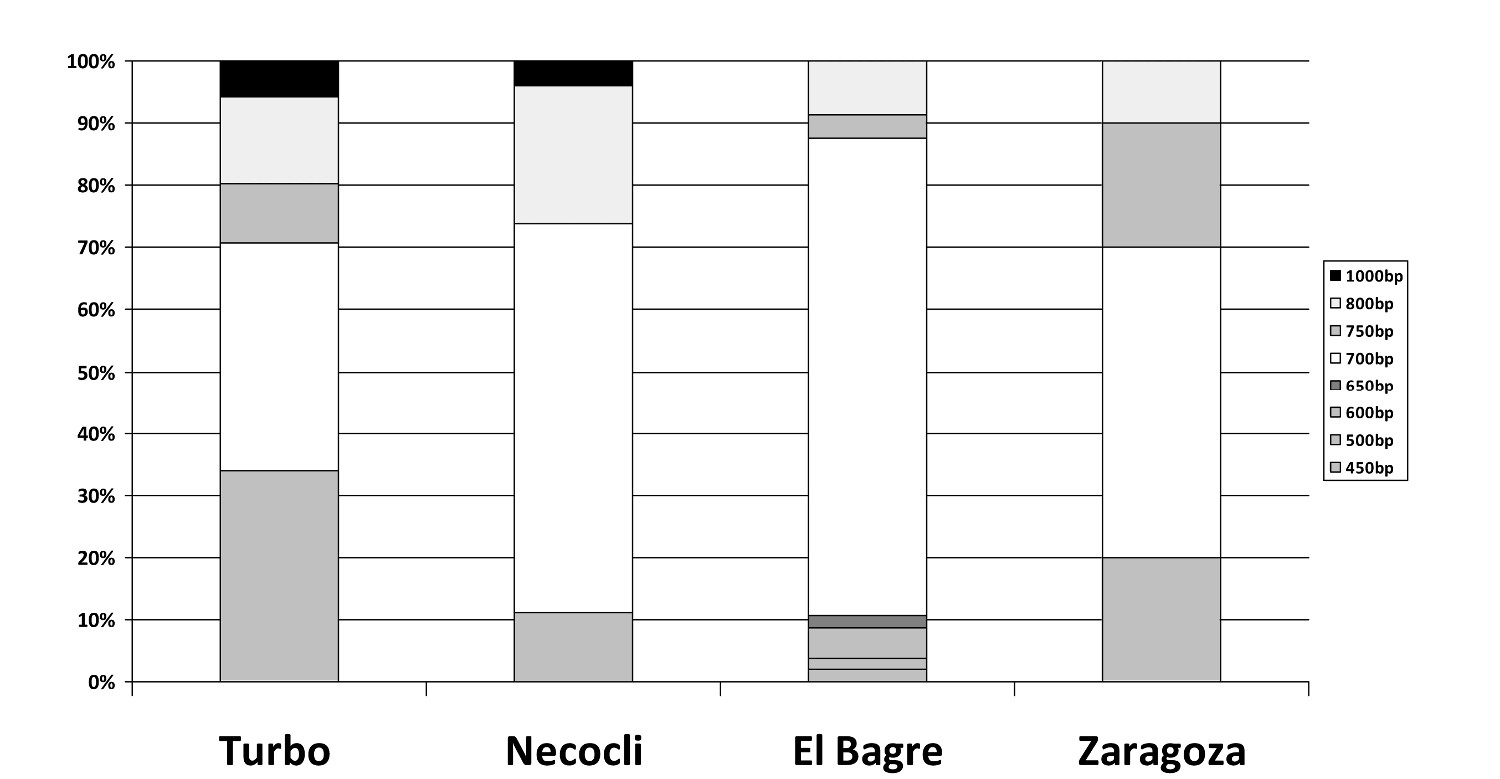
The frequencies of the individual glurp genotypes were high. In total, eight different genotypes were detected. The allelic variants ranged between 450 and 1,000 bp, and most samples exhibited the 700 bp sized fragment (58.1%), followed by the 600 (16.2%) and 800 (14.5%) bp fragments (Fig. 1).
Figure 1
Detail of the polymorphism size of the glurp gene in each locality. Frequencies for the amplified fragments, according to the size of the are presented for each locality.
Mean multiplicity of infection (MOI), calculated using data from each of the three marker genes, was 0.023 for msp1, 0.027 for msp2, and 0.047 for glurp.
A total 33 haplotypes were detected with a predominance of exclusive haplotypes in each of the localities studied. The highest number of exclusive haplotypes was observed in Necocli (n= 11), and the lowest number was detected in Turbo (n= 3). Shared haplotypes in the two main regions (Uraba and Bajo Cauca) were frequently found (Fig. 2).
Discussion
Polymorphic regions of the P. falciparum genes msp1, msp2 and glurp loci have been selected as the recommended markers for parasite genotyping in anti-malarial drug trials and efficacy studies 4 . However, the parasites' genetic profile has not been systematically documented in Colombia. In this study, a large number of archived P. falciparum positive pre-treatment parasites were genotyped in order to compare the diversity and allelic frequencies for these in two regions of Colombia exhibiting the highest levels of endemicity.
Homogeneity of P. falciparum populations was confirmed in these regions of Colombia, according to the genes studied. This is in striking contrast to observations made in the parasites from Africa where msp1, msp2 and glurp markers were highly polymorphic and had low allele frequencies 4 . These observations have been consistent overtime, as confirmed by diverse studies performed in the region from the works studies of 2000-2001 20 . In Table 1, a comparison is made of the different studies reported with the samples taken in Colombia. This illustrates an apparent reduced polymorphism based on an analysis of these genes, regardless of the diverse P. falciparum endemicity 21 , 22 . Homogenous P. falciparum populations, according to msp1 and msp2 after fixation of MAD-20 alleles, have also been reported by other authors (Table 1) 22 , 23 . However, a recent emergence of K1 and RO33 alleles is evidenced from the current study. Several factors might account for this phenomenon, such as the introduction of new alleles after migration events and/or the parasite´s selection after the introduction of new treatment schemes. The latter appears to be the case in Colombia since from 2006.
Genetic diversity is suggested by the differences in the amplified fragment size of the alleles detected for msp1 and msp2 from the different studies carried out in Colombia. This finding can be interpreted as allele variation or allele evolution. However, the application of different methodologies across these studies makes comparisons difficult, as suggested by Färnert et al 24 . It is recommended that a unified system be used to explore variability in Colombian samples. In any case, continuous monitoring of the presentationof msp1 alleles is worthy of pursuit in order to confirm and early define changes in the population's structure.
For glurp, a trend of diagnosing larger number of alleles has been detected over time and this is in direct relationship with endemicity and the number of samples evaluated 21 . This was also confirmed in the present study. Nevertheless, the number of alleles detected by this marker remains significantly low compared to African parasites 4 . Furthermore, the frequency of multiplicity of infection using the selected marker genes was low when compared with the African parasites 4. However, in the present study, a high frequency of polyclonal infection using glurp was detected during 2004-2006. Thereafter, it returned to lower levels. This can be explained by particular climatic phenomena affecting mosquito populations which may select for particular parasitic genotypes and subsequently change the transmission pattern. Alternatively, this may be the result of a natural process that was halted by the introduction of new anti-malarial treatments.
Based on these results we conclude that an analysis of glurp might be used successfully to genotype parasitic populations during new studies in Colombia aimed at exploring parasite population dynamics and the in vivo efficacy of anti-malarials. In addition, analysis of msp1 and msp2 can also be of value to make better comparisons with past studies, but not when the objective is to study parasites obtained from the same patient in a reduced period of time; for example, during treatment efficacy studies. Sequencing analysis of these genes in Colombian samples might prove useful to understand population dynamics in the different regions of the country and to further confirm the homogeneity of the populations.