Detection of genes mutations in the K-ras, H-ras and EGFR in samples of blood plasma and cervical smears for patients with cervical intraepithelial neoplasia III and cervical cancer
Dabeiba Adriana García, MSc1, Yazmín Rocío Arias, MSc2, Fabio Ancízar Aristizábal, PhD2
* Project supported by Universidad Nacional de Colombia, Bogotá, Colombia.
1. Pontificia Universidad Javeriana, Centro de Investigaciones Odontológicas, Bogotá, Colombia.
e-mail: garciad@javeriana.edu.co yrariasm@unal.edu.co
2. Universidad Nacional de Colombia, Departamento de Farmacia, Bogotá, Colombia. e-mail: faaristizabalg@unal.edu.co
Received for publication March 31, 2008 Accepted for publication January 15th, 2009
SUMMARY
Introduction: Cervical cancer is the second most important cancer in women worldwide, and the second cause of cancer death in women. It has been shown that the process of cervical carcinogenesis presents as genetic and epigenetic components as environmental issues. At present, many studies are addressed in searching for molecular markers such as mutations in oncogenes and/or tumor suppressor genes that are associated with the progression of this disease, the most studied candidate genes in cervical cancer in different populations have been H-ras, K-ras, EGFR among others.
Objective: The present study identified human papilloma virus (HPV) generic and specific in DNA-free plasma and cervical smears of invasive cervical cancer patients and patients with cervical intraepithelial neoplasia (CIN) III in addition to assessing genetic alterations, such as mutations in the genes H-ras, EGFR and K-ras.
Methods: To do so generic HPV was detected by PCR with primers GP5+/GP6+, and specific HPV 16 and 18 in E6/E7 region; to detect mutations in codon 12 of H-ras, codons 12 and 13 of K-ras and EGFR exon 21 was conducted by direct sequencing of PCR products of these gene fragments.
Results: Getting a good correlation between samples of blood plasma and cervical smears for both; the findings of HPV p=0.0374 and evaluated mutations p=0. In general, for EGFR in exon 21 mutations were not found, as for codons 12 and 13 in K-ras and codon 12 in H-ras.
Conclusion: The use of DNA in plasma may be relevant to the analysis of mutations and the presences of tumor markers are not available from other samples.
Keywords: Mutation; Cervical cancer; High-grade lesion; Human papillomavirus; H-ras; K-ras; EGFR.
Detección de mutaciones en los genes K-ras, H-ras y EGFR en muestras de plasma sanguíneo y cepillado cervical de pacientes con neoplasia intraepitelial cervical (NIC) III y cáncer de cuello uterino
RESUMEN
Introducción: El cáncer cervical es el segundo cáncer más importante en mujeres a nivel mundial y la segunda causa de muerte en mujeres por cáncer. Se ha demostrado que el proceso de carcinogénesis cervical presenta componentes tanto genéticos, epigenéticos y medio ambientales. En la actualidad, muchos estudios se encaminan en la búsqueda de marcadores moleculares como mutaciones en oncogenes y/o genes tumor supresor que se asocien con la progresión de esta entidad. Los genes candidatos más estudiados en cáncer cervical en distintas poblaciones han sido H-ras, K-ras, EGFR entre otros.
Objetivos: Se identificó el virus de papiloma humano (VPH) genérico y específico en el ADN libre de plasma y de cepillado cervical de pacientes con cáncer cervical invasivo y con neoplasia intraepitelial cervical (NIC) III además de evaluar alteraciones genéticas, como mutaciones en los genes H-ras, K-ras y EGFR.
Metodología: Para ello se detectó el VPH genérico mediante PCR con los iniciadores GP5+/GP6+, y específico para VPH 16 y 18 en la región E6/E7. Para detectar las mutaciones en el codón 12 de H-ras, codones 12 y 13 de K-ras y el exón 21 de EGFR se realizó mediante secuenciación directa de los productos de PCR de estos fragmentos génicos.
Resultados: Obteniendo una buena correlación entre las muestras de plasma sanguíneo y los cepillados cervicales, tanto para los hallazgos de VPH p=0.0374 como para las mutaciones evaluadas p=0. En general, para EGFR en el exón 21 no se encontraron mutaciones, al igual que para los codones 12 y 13 en K-ras y codón 12 en H-ras.
Conclusión: El uso del ADN presente en el plasma puede ser relevante para el análisis de mutaciones y de la presencia de marcadores tumorales cuando no se dispone de otras muestras.
Palabras clave: Mutaciones; Cáncer cervical; Lesión intraepitelial de alto grado; Papilomavirus humano; H-ras; K-ras; EGFR.
Cervical cancer is one of the world most important diseases. In Colombia, estimated incidence is 29.4 cases per every 100,000 inhabitants1. Cervical carcinogenesis begins with non invasive squamous lesions that progress to low and high grade intraepithelial squamous lesions; and eventually generate an invasive carcinoma2. However, predicting which of them finally progress and which will reverse is currently impossible as the entire cellular and molecular mechanisms involved in progression are unknown. Even more, in spite of recent advances uncovering several routes leading to cervical carcinogenesis, no molecular marker been closely enough associated with progression or clinical outcome has been identified to date.
Multiple findings provide evidence about the increased presence of cellular and viral DNA in plasma from cervical cancer patients3. Patients with various cancers like colon, esophagus, breast, pancreas, head and neck and liver present increased levels of free circulating plasma DNA. In addition to this, many genetic (such as amplified oncogenes, punctual mutations involving Tumor Suppressor Gene, microsatellite instabilities, etc) or epygenetic features (such as TSG methylated promoters) resemble those detected directly in the corresponding tumors4. So, plasma DNA from cancer patients becomes a promising target for detecting molecular cancer associated alterations which could be used as early cancer markers or an ongoing relapse of the disease after the primary treatment. In cervical cancer, mutations in EGFR, H-ras and K-ras, are some of the most frequent genetic changes studied.
H-ras (Harvey) and K-ras (Kirsten) are located on chromosomes 11 and 12 respectively. These oncogenes code a 21 kDa protein called p21 which exhibits GTPase activity and is implied in cell cycle regulation5. Constitutive activation of them is mainly caused by punctual mutations at codons 12, 13 and 61, leading to the inhibition of its GTPase activity. High levels of ras function have been demonstrated in lung, bladder, breast, colon and cervical cancer among others. Besides, such mutations can be found in normal looking tissue and pre neoplastic lesions from cervical cancer patients, suggesting an important role in carcinogenesis6.
EGFR (ErbB1) codes for a 170 kDa membrane receptor involved in cell proliferation. It is located in chromosome 7 and belongs to the ErbB tyrosine kinase family of receptors. Other members are Her2/neu (ErbB 2), ErbB 3 and ErbB 4. Among the extracellular ligands of EGFR are the EGF and the TGFa7. Ligand binding induces self phosphorylation and dimerization of the receptor with some conformational changes associated which initiate transduction of a transforming, anti-apoptotic, proliferative signal. In addition, mutations at exons 18-24, known to alterate its tyrosine kinase activity, have been found in epithelial tumors such as those on breast and cervix7.
Since mutations in K-ras and H-ras GTPase domains have been presented as promising progression associated molecular markers, and so potential tools for relapse/metastasis detection in solid tumors, we decided to search whether mutations found in cervical scrapes from CIN III and cervical cancer patients in genes these loci are accurately resembled by alterations in free circulating plasma DNA from themselves. Presence of HPV was also evaluated in both samples.
MATERIALS AND METHODS
Patients and sampling procedure. This is a descriptive, crossectional study involving patients conveniently chosen during a 6 months lapse at a third level hospital in Bogotá DC. A cervical scrape and a peripheral blood sample were obtained from 26 patients who had been diagnosed with either a high grade intraepithelial lesion (HSIL) or cervical cancer (CC), during confirmative colposcopy. All of the patients were fully informed and required to sign a written consent before being enrolled in the study. None of them had received any therapy at the time of sampling.
When obtained, every sample obtained by cervical scrape was introduced into a tube with Timerosal 0.05% in PBS. The tubes were kept on ice until their arrival to lab for processing. Each blood sample was drained into an EDTA added Venoject® tubes by venopuncture.
Upon arrival, each cell suspension was centrifuged at 3000 g for 10 minutes, then the supernatant was discarded and de pellet was suspended in 1 ml of 10 mM Tris-HCl buffer (pH 8.3) and boiled during 10 minutes. After a one minute 3,000 g centrifugation the supernatant was directly used as PCR template source. Blood samples were centrifuged 20 minutes at 3,000 g, the upper plasma face was separated and further centrifuged at 8000 g for 3 minutes to completely deplete cellular debris. DNA extraction was carried out with the QIAamp® DNA Mini Kit (QIAGEN; Germany) following manufactures instructions. DNA isolates were then subjected to a pre-PCR «restoration» treatment as previously described8. Briefly, each plasma DNA sample is incubated at 55°C with dNTPs 200 mM for 1 hour. The treatment is performed in the presence of Tris-HCl 10 mM, 50 mM KCl, 0.1% triton X-100, and MgCl2 3 mM. Then one unit of Taq DNA polymerase is added an incubated 20 minutes more at 72°C.
DNA quality. Since DNA in plasma is subjected to multiple degradative conditions it is necessary to that DNA extracted is quality enough to support PCR. Quality of the DNA from cervical scrapes and plasma (restored and non restored) was evaluated by amplification of the b-globin gene (HHB) with primers PCO3 (ACACAACTGTGTTCACTAGC) and PCO4 (CAA CTTCATCCACCTTCACC) with Tris-HCl 10 mM, 50 mM KCl, 0.1% triton X-100, MgCl2 3 mM, 7.5 pmol each primer, 1U Taq DNA pol and 200 mM each dNTP. Reactions were performed in a MyCyclerTMtermocycler (BioRad Laboratorios, USA) by running the following cycling program: 95°C - 3 minutes, and 95°C - 20 seconds, 58°C - 20 seconds and 72°C - 20 seconds for 40 repetitions. Products were analyzed by 2% agarose electrophoresis where expected product is 110 bp long. Lymphocyte DNA was used as positive control.
HPV detection and typification. Generic detection of HPV was also performed by PCR with primers GP5+ (TTTGTTACTGTGGTAGATACTA) and GP6+ (GAAAAATAAACTGTAAATCATA). Reagent conditions were as follows: Tris-HCl 10mM, 50 mM KCl, 0.1% triton X-100, 3 mM MgCl2, 0.3 mM each primer, 1U Taq DNA pol and 200 uM each dNTP; with the following program: 95°C - 4 minutes; and 95°C - 1 minute, 47°C - 1 minute and 72°C - 1.5 minutes for 40 repetitions. Products were equally analyzed by electrophoresis as described above. DNA isolates from both cervical scrape and restored plasma DNA from HPV positive cases were further assayed for HPV16 and 18 by PCR targeted to the E6/E7 region. Primers used were as follows: HPV16, primers ATCATCAAG AACACGTAGAG and GATCAGTTGTCTCTGGTT GCAAAT; HPV18, primers GATTTCACAACATA GCTGGG and TGCCTTAGGTCCATGCATAC. In both cases the first is the sense primer while the second is the antisense primer. Reagent conditions were identical to those for generic HPV detection, while cycling conditions were: 94°C - 2 minutes; 94°C - 30 seconds, 60°C - 30 seconds y 72°C - 30 seconds. As before, products were analyzed by agarose electrophoresis. Final reaction volumes were 50 ml for generic detection and 25 ml for type specific detection. For all HPV detections plasmids pBR322 Zur Hausen donation containing either HPV16 or HPV18 complete genomes were used as positive controls.
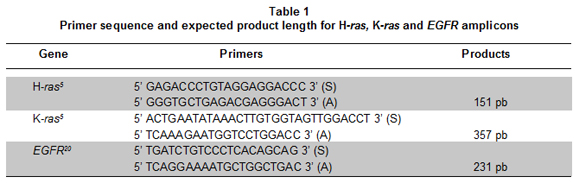
H-ras, K-ras and EGFR mutation analysis. PCR products were also generated to verify base identities at codon 12 of H-ras, codons 12 and 13 of K-ras, and exon 21 of EGFR from cervical scrapes and restored plasma DNAs. Primers used are showed in Table 1. Reagent conditions were as used in assays above, and a final volume of 25 ml was used. Cycling conditions were: an initial 94°C - 5 min denaturation step for all the amplicons; 94°C - 30 sec, 60°C - 30 sec and 72°C - 30 sec for H-ras and K-ras; while 94°C - 30 sec, 58°C - 30 sec and 72°C - 30 sec was used for EGFR. Products were purified with the Wizard® DNA Clean-Up System (Promega) according to manufacturer instructions, and sequenced in an ABI 3730 XL sequencer (Applied Biosystems, Foster City, CA) by McLabs in San Francisco CA.
Ethical issues. This study was respected the ethic and legal aspects for in humans research stipulated in Resolution 008430 of 1993 (Colombian Ministry of Health), and the 2004 modified Helsinki declaration (1975). The patients enrolled in this study voluntarily accepted to participate after reading and understanding the consent form. This form was approved by the Ethics committees of the Javeriana and National Universities (Bogotá).
Statistical analysis. A data base was constructed with patient’s information in an EXCEL file (Microsoft). A one variable descriptive analysis was performed with qualitative variables like the presence of HPV and the presence of mutations in the sequences. Frequency distribution was determined for each one, and association tests were applied (Pearson’s c2) to link cervical scrape and plasma DNA isolates.
RESULTS
HHB (b-globin) amplification confirmed that DNA isolates from cervical scrapes and plasma were quality enough to generate reliable and reproducible PCR amplifications. Besides, the pre-PCR restoration treatment demonstrated to improve template quality, allowing a more sensitive detection of this single copy gene (Figure 1). In gel measurement of the band relative optical density (ROD), shows a great increase for HHB amplification upon plasma DNA restoration. Initial average ROD was 427.2 while the same parameter raised to 1032.2 after the treatment. Measures showed 286.3 and 183.4 as standard deviation respectively.
100% of cervical scrapes and 84.6% of plasmas were positive for HPV, generic detection targets the L1 region of viral genome coding for the viral capsomer. The stretch amplified is a 142 bp long amplicon (Table 2). Detection of this amplicon in cervical scrapes and plasma showed great correlation as verified by a P value of 0.0374.
Specific subtype detection in cervical scrapes revealed 61.5% and 11.5% of incidence for HPV16 and 18 respectively among HPV positive cases (Table 3). HPV16 was demonstrated in 50% of the plasmas while HPV18 was detected in 11.5% of them (Table 4). In two patients, who presented both cervical cancer and a CIN III lesion at colposcopy, subtype specific HPV detection was not possible. In general, though HPV16 detection in cervical scrapes was not fully matched to that in plasmas, results demonstrate concordance (61.4% and 50% respectively). No plasma sample showed a HPV subtype not found in the corresponding cervical scrape. On the other hand, HPV18 showed equal incidence in scrapes and plasmas (11.5%).
K-ras, H-ras and EGFR were successfully amplified in both cervical scrapes and plasma DNA from all the cases (Figure 2). Isoform A of EGFR was the sequence found in all the patients. Exon 1 was shown not to be altered in any of the cases. Codon 12 of H-Ras didn’t show mutations either; however, two patients showed a third base change at codon 27 (CAT changed to CAC) detected in both cervical scrape and matched plasma DNA. Since it is a silent mutation, it continues to code for Histidine. Besides, no mutations were seen in codons 12 and 13 of K-ras; however, all the patients showed a CCT sequence in codon 11 which codes for Proline, instead of GCT corresponding to Alanine as occurs in the type sequence. Even though both amino acids are non polar, they greatly differ in the contribution they do to higher order protein holding (Figure 3).
DISCUSSION
Both genomic and viral DNA can be effectively retrieved from plasma from CIN III and invasive CC patients, confirming plasma analysis as an important tool for diagnosis and prognosis3. However, this material could have suffered single and double strand breakage reducing its quality as PCR template. The restoration treatment applied to plasma DNA isolates allows single strand gaps to be filled by using the information of the complementary strand as template, so improving its quality for genetic markers detection. Since the pre-PCR restoration treatment used here does not includes the repetitive synthesis cycles proper of PCR it does not increases the number of copies of the target sequence. The treatment only restores available copies so that primers find them more efficiently8.
Regarding HPV detection in cervical scrapes, it was 100% incident among the cases analyzed; this is compatible with the very high association commonly found and supports the causal relationship between HPV and cervical neoplasia9. HPV detection in plasma from CIN III and CC patients allowed to classify 84.6% of the cases as positive, which demonstrate a better detection rate than that presented previously studies by Wei et al.4and Widschwendter et al.10where HPV was confirmed in 64.7% and 45% of plasmas from patients with positive detection in tumor respectively. Our better detection rate can be explained by better template integrity after the restoration treatment applied.
HPV16 was detected in 61.54% of cases (cervical scrape detection), resembling previous studies by Rabelo-Santos et al.11in Brazil (57.1%); on the other hand, HPV18 was found in 11.5% of our cases, similarly to detection rates presented by Bosch et al.12and Muñoz13, whose detection rates were 14% and 13.6% respectively. Specific detection in plasma allowed HPV16 detection in 50% of cases, while HPV18 was found in 11.5% of them. Previous data from assays targeted to plasma indicate a very low rate even for generic detection. Specific detection and typification have been consequently very limited. As an example, results presented by Dong3were 3.5% for subtype 16 and 2.1% for HPV18.
EGFR gene was found unaltered at exon 21 in all our cases. Similar results are reported by Pühringer-Oppermann et al.14 for esophageal cancer. Sequence mutations in codon 12 of H-ras were not found in our cases either resembling reports by Mammas et al.5in cervical cancer, and Vachtenheim et al.15 for lung squamous carcinomas; however, two of the cases showed a mutation at codon 27 (CAT to CAC) both in scrapes and matched plasmas. Similar results are presented by Lev et al.16in melanoma cells.
K-ras, was not found mutated at codons 12 and 13 either, as reported by Pochylski and Kwasniewska17in cervical squamous cell carcinomas. However, codon 11 was found to be changed from GCT to CCT (Ala to Pro) in all the cases. This mutation was previously found by Gu et al.18and Hongyo et al.19in patients with lung carcinomas and gastric adenomas respectively. In general, a good correlation was observed between sequences of H-ras, K-ras and EGFR found in cervical scrapes and plasmas. As also demonstrated by Dong, et al.3, sequence alterations at specific loci found in plasma DNA isolates from cancer patients closely resemble those present in tumors.
We conclude that analyzing free circulating DNA in plasma from cancer patients is a reliable approximation to tumor genetic features, allowing the analysis of numerous genetic markers when obtaining tumor mass samples is not an option.
ACKNOWLEDGEMENTS
The authors thank the Departamento de Farmacia and the Instituto de Biotecnología of the Universidad Nacional de Colombia (Bogotá, Colombia). The DIB (División de Investigaciones Bogotá) of the Universidad Nacional provided all the funds to afford this project (Code: 2010000). We thank the great help received from doctor Antonio Huertas Salgado (Instituto Nacional de Cancerología ESE, Bogotá) at sequence analysis, and doctor Fabián Carrillo for their assistance in translation into English of this article.
REFERENCES
1. Ferlay J, Bray F, Pisani P, Parkin DM. GLOBOCAN 2002. Cancer incidence, mortality and prevalence worldwide (IARC). Cancer Base N° 5, version 2.0. Lyon: IARC Press; 2004.
2. Song SH, Lee JK, Oh MJ, Hur JY, Park YK, Saw HSssRisk factors for the progression or persistence of untreated mild dysplasia of the uterine cervix.Int J Gynecol Cancer. 2006; 16: 1608-13.
3. Dong SM, Pai PI, Rha SH, Hildesheim A, Kurman RJ, Schwartz PE, et al. Detection and quantitation of human papillomavirus DNA in the plasma of patients with cervical carcinoma. Cancer Epidemiol Biomarkers Prev. 2002; 11: 3-6.
4. Wei YC, Chou YS, Chu TY. Detection and typing of minimal human popillomavirus DNA in plasma. Int J Gynaecol Obstet. 2007; 96: 112-6.
5. Mammas IN, Zafiropoulos A, Koumantakis E, Sifakis S, Spandidos DA. Transcriptional activation of H- and N-ras oncogenes in human cervical cancer. Gynecol Oncol. 2004; 92: 941-8.
6. Lee JH, Lee SK, Yang MH, Ahmed MM, Mohiuddin M, Lee EY. Expression and mutation of H-ras in uterine cervical cancer. Gynecol Oncol. 1996; 62: 49-54.
7. Kim YT, Park SW, Kim JW. Correlation between expression of EGFR and the prognosis of patients with cervical carcinoma. Gynecol Oncol. 2002; 87: 84-9.
8. Arias YR, Carrillo EF, Aristizábal FA. Plasma DNA restoration for PCR applications. J Clin Pathol. 2007; 60: 952-4.
9. Bosch F, Lorincz A, Muñoz N, Meijer CJLM, Shah KV. The causal relation between human papillomavirus and cervical cancer. J Clin Pathol. 2002; 55: 244-65.
10. Widschwendter A, Blassnig A, Wiedemair A, Müller-Holzner E, Müller HM, Marth C. Human papillomavirus DNA in sera of cervical cancer patients as tumor marker. Cancer Lett. 2003; 202: 231-9.
11. Rabelo-Santos SH, Zeferino L, Villa LL, Sobrinho JP, Amaral RG, Magalhães AV. Human papillomavirus prevalence among women with cervical intraepithelial neoplasia III and invasive cervical cancer from Goiânia, Brazil. Mem Inst Oswaldo Cruz. 2003; 98: 181-4.
12. Bosch FX, Manos MM, Muñoz N, Sherman M, Jansen AM, Peto J, et al. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. International Biological Study on Cervical Cancer (IBSCC) Study Group. J Natl Cancer Inst. 1995; 87: 796-802.
13. Muñoz N, Méndez F, Posso H, Molano M, van den Brule AJ, Ronderos M, et al. Incidence, duration, and determinants of cervical human papillomavirus infection in a cohort of Colombian women with normal cytological results. J Infect Dis. 2004; 190: 2077-87.
14.Pühringer-Oppermann FA, Stein HJ, Sarbia M. Lack of EGFR gene mutations in exons 19 and 21 in esophageal (Barrett’s) adenocarcinomas. Dis Esophagus. 2007; 20: 9-11.
15. Vachtenheim J, Horakova I, Novotna H, Opaalka P, Roubkova H. Mutations of K-ras oncogene and absence of H-ras mutations in squamous cell carcinomas of the lung. Clin Cancer Res. 1995; 1: 359-65.
16. Lev DC, Onn A, Melinkova VO, Miller C, Stone V, Ruiz M, et al. Exposure of melanoma cells to dacarbazine results in Enhanced tumor growth and metastasis in vivo. J Clin Oncol. 2004; 22: 2092-100.
17. Pochylski T, Kwasniewska A. Absence of point mutation in codons 12 and 13 of K-ras oncogene in HPV-associated high grade dysplasia and squamous cell cervical carcinoma. Eur J Obst Gynecol Rep Biol. 2003; 111: 68-73.
18. Gu Q, Li L, Zhang Y, Li X, Xiao Y, Wu X. Sequencing analysis of human K-ras oncogene exon 1 in Chinese people. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 1998; 15: 98-100.
19. Hongyo T, Buzard GS, Palli D, Weghorst CM, Amorosi A, Galli M, et al. Mutations of the K-ras and p53 genes in gastric adenocarcinomas from a high-incidence region around Florence, Italy. Cancer Res. 1995; 55: 2665-72.
20. Moroni M, Veronese S, Benvenuti S, Marrapese G, Sartore-Bianchi A, Di Nicolantonio F, et al. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: a cohort study. Lancet Oncol. 2005; 6: 279-86.