Design of a molecular method for subspecies specific identification of Klebsiella pneumoniae by using the 16S ribosomal subunit gene
Nelson Enrique Arenas, Msc1, Juan Carlos Polanco, PhD2, Sandra Milena Coronado, MSc1, Clara Juliana Durango, Quim1, Arley Gómez, PhD1,3*
1. Grupo de Investigación en Ciencias Biomédicas, Centro de Investigaciones Biomédicas. Facultad de Ciencias de la Salud, Universidad del Quindío, Armenia, Colombia. e-mail: nearenass@unal.edu.co arley.gomez24@urosario.edu.co sandracoronado360@yahoo.com clara_juliana_d@yahoo.com.br
2. División de Genética Molecular y Biología del Desarrollo, Instituto de Biociencias Moleculares, Universidad de Queensland, Brisbane, Australia. polancojc@yahoo.com
3. Director, Unidad de Medicina Tropical y Enfermedades Infecciosas, Universidad del Rosario, Bogotá, DC, Colombia.
Received for publication February 12, 2008 Accepted for publication April 14, 2009
SUMMARY
Introduction: Rhinoscleroma is caused by Klebsiella pneumoniae rhinoscleromatis and the ozena infections caused by K. pneumoniae ozaenae, both infections affect the upper respiratory tract. In the first clinical phases the symptoms are unspecific, and the disease can be misdiagnosed as a common cold, therefore antimicrobial therapy cannot reach effective results and patients must be following up for several years since the infection became chronic.
Objective: To identify Klebsiella subspecies using a specific assay based on amplicons restriction of a gene which encodes 16S subunit ribosomal (rDNA16S).
Methodology: Specific restriction patterns were generated; using reported sequences from rDNA16S gene and bioinformatics programs MACAW, PFE, GENEDOC and GENE RUNNER. Amplification and restriction assays were standardized.
Results: Predictions in silico allowed to propose an algorithm for Klebsiella species and subspecies identification. Two reference strains were included and two clinical isolates which were biotyped and identified by the proposed method. rDNA16S gene restriction patterns showed differences regarding the initially identified species for conventional methods. Additionally two patterns of bands were observed for K. pneumoniae rhinoscleromatis, indicating the polymorphisms presence in the rDNA16S gene.
Conclusions: It was confirmed the difficulty to identify K. pneumoniae subspecies by conventional methods. Implementation of this technique could allow an accurate and rapid differentiation among K. pneumoniae ozaenae and K. pneumoniae rhinoscleromatis aetiological agents of two frequently misdiagnosed infections. Antimicrobial therapy usually could be ineffective, especially in chronic patients. Finally it is considered very important to enlarge the study by using more clinical and reference strains.
Keywords: Klebsiella pneumoniae rhinoscleromatis; K. pneumoniae ozaenae; K. pneumoniae pneumoniae; Diagnosis.
Diseño de un método molecular para la identificación específica de Klebsiella pneumoniae a nivel de subespecie, usando el gen que codifica para la subunidad ribosomal 16S
RESUMEN
Introducción: El rinoescleroma es causado por Klebsiella pneumoniae rhinoscleromatis y la ocena por Klebsiella pneumoniae ozaenae respectivamente. Estas infecciones se presentan sobre todo en el tracto respiratorio superior y tienen una sintomatología inespecífica en sus fases iniciales por lo cual se pueden confundir con el catarro común. Las dificultades de establecer un diagnóstico oportuno tienen repercusiones negativas en la terapia antimicrobiana, porque puede no ser efectiva y hacer que la enfermedad evolucione a una fase crónica cuyo seguimiento puede implicar muchos años.
Objetivo: Diseñar un ensayo molecular para la identificación a nivel de subespecie de bacterias del género Klebsiella basado en restricción de amplicones del gen que codifica para la subunidad ribosomal 16S (ADNr 16S).
Metodología: Se generaron patrones de restricción específicos, utilizando secuencias informadas del gen ADNr 16S y los programas bioinformáticos MACAW, PFE, GENEDOC y GENE RUNNER. Se estandarizaron las condiciones para la amplificación y restricción para el ensayo experimental.
Resultados: Las predicciones in silico permitieron proponer un algoritmo para la identificación a nivel de especie y subespecie de las especies del género Klebsiella. Se incluyeron dos cepas de referencia y dos aislados clínicos, que se biotipificaron e identificaron por el método propuesto; los patrones de restricción obtenidos del gen ADNr 16S evidenciaron diferencias con respecto a la especie inicialmente identificada por métodos convencionales. Además se encontraron dos patrones de bandas en Klebsiella pneumoniae rhinoscleromatis, indicando la presencia de polimorfismos en el gen ADNr 16S para esta subespecie.
Conclusiones: Se confirmó la dificultad para identificar Klebsiella pneumoniae a nivel de subespecie por métodos convencionales. La implementación de esta técnica podría permitir la diferenciación temprana entre Klebsiella pneumoniae ozaenae y Klebsiella pneumoniae rhinoscleromatis que causan dos infecciones tratadas por lo general de forma empírica y como consecuencia de esto, la terapia antimicrobiana suele no ser efectiva, en especial en pacientes crónicos. Se requiere ampliar los estudios con un número mayor de cepas de referencia y aislados clínicos.
Palabras clave: Klebsiella pneumoniae rhinoscleromatis; Klebsiella pneumoniae ozaenae; Klebsiella pneumoniae pneumoniae; Diagnóstico.
Historically, taxonomic position of Klebsiella genus has been highlighted by several reclassifications and emendations, therefore connoted with controversial framework. Actually recognized Klebsiella species include K. pneumoniae with three subspecies (K. pneumoniae pneumoniae, K. pneumoniae rhinoscleromatis and K. pneumoniae ozaenae), K. oxytoca (with two subgroups), K. variicola and also coming K. granulomatis (formally named Calymmatobacterium granulomatis)1.
A further classification system recognizes other species as K. planticola and K. terrigena isolated form environmental sources, K. ornithinolytica and Enterobacter aerogenes, which has been named as K. mobilis and is a genus related specie2.
Recently these environmental species have been transferred to a new genus called Raoultella, which was recognized with basis to differences through meanly by comparative analysis of ribosomal 16S genes (rDNA 16S) and the ARN polymerase beta subunit (RpoB) from recognized species of Klebsiella genus and related enterobacteria, as consequence they constitute two distant lineages on phylogenetic studies3.
On the other hand, several classification systems based on Klebsiella infections has been proposed and thereby international nomenclature has changed and taxon identification could be cumbersome, especially for environmental species2.
Traditionally laboratories carry out Klebsiella differentiation at species and subspecies level by conventional methods; however, these techniques lack of specificity, reproducibility and sometimes there is no correlation when comparison is made with commercial methods such as API 20E system. This situation has been associated with the high similarity and homology on Klebsiella genome which make more complex the differentiation with specific genetic markers of clinical isolates and environmental strains4.
Usually at genus level K. pneumoniae, is considered the most clinical important specie, since it’s responsible of nosocomial and community outbreaks and can generate a wide range of infections due to bacterial ability to colonize and spread out on gastrointestinal, urinary and respiratory tracts5.
Two K. pneumoniae subspecies produce specific respiratory infections. K. pneumoniae rhinoescleromatis is the etiological agent of rhinoscleroma or scleroma infection which is characterized by a granulomatous and chronic process of insidious evolution that affects the mucosa from the upper respiratory tract and might lead to bone invasion and airway obstruction. Clinically, three phases are recognized: catarral-athrophic, granulomatous and cicatricial6.
Klebsiella pneumoniae ozaenae produce ozena or atrophic rhinitis, an infection compromising nasal epithelium which results in clinic symptoms such as: anosmia, green mucopurulent and fetid exudates, and nose obstruction whose histological findings are consistent with focal areas of squamous metaplastic damage of the mucosal glands and therefore mucus secretion alteration7.
Rhinoscleroma and ozena infections are frequently misdiagnosed as a common cold and sinusitis on the initial phases, due to the vague signs and symptoms. These infections show low frequency and commonly the diagnoses are done in a late stage, therapeutic choice is difficult and the results are not fully effective, especially for patients with chronic infection.
The evolution of these infections represents a serious threat for the patient, because of chronic tissue damage and sequelae, recurrences are common events associated with patient infected with resistant strains, which reduces treatment efficacy, increases the economic cost and the patient should be followed up for several years8.
Available methods for K. pneumoniae strains identification allow a much better differentiation when used in combination but are more expensive, laborious and not accurate enough for epidemiological studies because of their low discriminative power and reproducibility2.
Molecular techniques are a useful tool for infectious diseases diagnosis; the development of polymerase chain reaction (PCR) based systems have been implemented successfully by their reproducibility, accuracy and specificity for specific recognition of minimal differences at genotypic level on highly related organisms and to overcome the conventional test limitations, due to their dependence of physiologic and metabolic activity.
This study, designed a molecular assay based on PRA (PCR Restriction Assay) of rDNA16S gene from K. pneumoniae for differentiation at the subspecies level.
MATERIALS AND METHODS
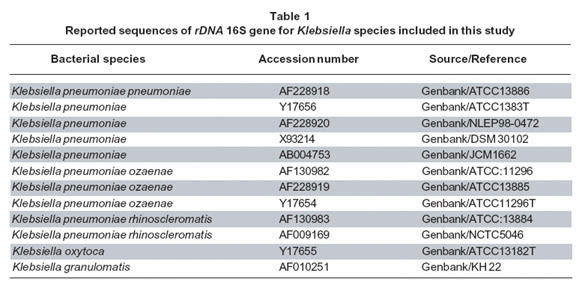
Bioinformatic analysis. Reported sequences for the rDNA 16S genes of Klebsiella genus were obtained from the Genbank and some described sequences for setting phylogenetic relationships of related Enterobacteriacea species were also included9 (Table 1). Multiples alignments were performed with MACAW version 2.0.5; manually edited on PFE software and imported to GENEDOC program for restriction patterns prediction based on rDNA 16S sequence alignments. Primers were designed on GENE RUNNER program. Primer specificity and universality evaluation was carried out by using BLASTn program10.
Phenotypic identification. Two K. pneumoniae reference strains (K. pneumoniae ozaenae PUJ 040 and K. pneumoniae PUJ 017) were obtained from the Microorganism Collection from the Pontificia Universidad Javeriana (recognized by World Federation for Culture Collections) and two clinical isolates (referred as UQ001 y UQ002). MacConkey agar (BD) was used for primary culture. Conventional test were performed by bacterial culture on: Indole (BD), Citrate Simmons (Oxoid), Voges Proskauer (BD), Triple Sugar-Iron (BD), Urea (BD), Motility, Methyl Red (Oxoid) and gas production (BD)11. Reference strains were typed with API 20E commercial system (BioMèrieux, Marcy l‘Etoile, France).
Genotypic methods. DNA genomic extraction was carried out according to standardized protocol for aerobic bacteria12. rDNA 16S gene amplification from Klebsiella genus species were performed under the following reaction conditions: Klebrib-1 (5’-GTAATGTCTGGG AAACTGCC-3’) [0.5µM] and Klebrib-2A (3’- CCACC TTCCTCCAGTTTATC-5’) [0.5µM] Taq polymerase [0.25U], MgCl [1.5 mM], dNTPs [0.2 mM], PCR buffer [1X] and DNA sample [10 ng/µl], adjusted to a final volume of 50 µl. Amplification conditions were 94°C for 1 min; 40 cycles of 94°C for 1 min, 58°C for 1 min, and 72°C for 1 min; and a final step at 72°C for 3 min. PCR product (1069 bp) was purified with Wizard Genomics (DNA Prep., Promega) using the supplier’s recommended conditions. Restriction enzyme digestion was performed using Sal I, Ban II (37ºC) and Taq I (65ºC) enzymes, each reaction was performed either with 17 µl rDNA, 2 µl Multi Core buffer and 1 µl enzyme restriction (10 U/ µl) to 20 µl final volume. Every solution was mixed and incubated to optimum temperature of each restriction enzyme during 2 hours. Restriction reaction was inactivated with 5 µl EDTA [0.5 M] for 15 minutes. Analysis of the band patterns was obtained by visual interpretation on ethidium bromide stained electrophoresis gels [Agarose 3%].
RESULTS
In silico design of molecular technique. Alignments obtained with reported rDNA 16S sequences (Genbank) of Klebsiella species showed a high gene similarity interspecifically and intraspecifically. A simple algorithm for specific differentiation at subspecies level with single rDNA 16S gene digestions for K. pneumoniae ozaenae, K. pneumoniae rhinoscleromatis and K. pneumoniae pneumoniae was generated. Ban II enzyme generated an 816 bp specific fragment for the differentiation of K. pneumoniae ozaenae (Figure 1A).
Specific discrimination for K. pneumoniae rhinoscleromatis was tested in silico through gene pattern prediction of 759 bp and 311 bp of a sequence rDNA 16S gene with Sal I restriction enzyme (Figure 2A); nevertheless, due to polymorphism described among the ADNr 16S gene for K. pneumoniae rhinoscleromatis, it was necessary to search an alternative enzyme which allows to differentiate a second gen isotype, thus Taq I enzyme generated exclusive band patterns based on polymorphic sequence (Figure 3A). Consequently, it was crucial to take into account this finding for experimental confirmation of predicted band patterns for K. pneumoniae rhinoscleromatis reference strains and clinical isolates.
Finally, on the basis of clinical significance for differentiation between K. pneumoniae and K. oxytoca, additional in silico prediction showed that Rsa I enzyme can produce 483 bp, 319 bp y 293 bp fragment restriction in comparison with 776 bp and 293 bp fragments predicted in the remaining species (Data not shown).
Phenotypic identification of studied strains. The results obtained by using conventional biochemical test confirmed the significant difficulties for Klebsiella differentiation at the species level, even when reference strains were included and biotyping by other methods; some inconsistent findings were obtained for Methyl red and Voges Proskauer reactions.
This result uncertainty did not allow us to define clearly the species identity according to fermentation profile observed on media culture and bacterial strains were characterized only at the genus level. Species confirmation for reference strains were performed by API 20E commercial system, showing that K. pneumoniae ozaenae (strain PUJ 040) generated a numeric pattern concordant with K. oxytoca. (Code 5255773), this divergence was based on the differences obtained from biochemical test: indole production, citrate Simmons, urea, Voges Proskauer and sucrose. K. pneumoniae (PUJ 017) identification was concordant with pneumoniae subspecies (code 1215773) according to supplier’s instructions.
Conditions standardization for rDNA 16S amplification and restriction. ADNr 16S gene generated an amplification product of 1070 pair bases according to the expected size (Figure 4).
Amplicon restriction with Ban II showed the absence of the 816 bp specific fragment for K. pneumoniae ozaenae (PUJ 040) even when it was included a reference strain for this subspecies; however, for the remaining strains a 500 bp, 310 bp y 250 bp pattern bands were observed, which was concordant with in silico prediction for K. pneumoniae subspecies (Figure 1B). Sal I enzyme did not produce any amplicon cuts for K. rhinoscleromatis and K. pneumoniae subspecies but for K. ozaenae subspecies two bands (760 bp and 310 bp approximately) were observed (Figure 2B).
Based on restriction enzyme Taq I, similar band patterns (520 bp, 360 bp, 220 bp and 190 bp approximately) were obtained for K. pneumoniae pneumoniae (PUJ 017) and for a clinical sample. The remaining strains produced a (740 bp and 220 bp bands) which allowed us to differentiate K. pneumoniae subspecies from the other strains. Although, we reported two different patterns obtained from in silico prediction only one band pattern for K. pneumonia rhinoscleromatis (520, 360, 220 and 190 bp approximately) was experimentally determined (Figure 3).
The proposed molecular assay start with rDNA 16S amplification on the clinical sample corresponding to Klebsiella genus; then an initial restriction with Ban II lead to K. pneumonia ozaenae identification, and afterwards Sal I or Taq I restriction could differentiate among K. pneumoniae rhinoscleromatis and K. pneumoniae pneumoniae (Figure 5).
In general, the proposed molecular assay allow to carry out consecutive digestions to 16S ribosomal gene Amplicon with low cost restriction enzymes commercially available; easy reading and interpretation due to a low bands number (patterns); and none special software is required for data analysis and finally the test implementation would facilitate diagnosis on labs equipped for simple tests of molecular biology.
DISCUSSION
Klebsiella pneumoniae rhinoscleromatis and K. pneumoniae ozaenae typing by conventional tests is based on phenotypic profile assignment which is compared with high concordance of a standard profile; nevertheless this method can lead to erroneous or incomplete identification due to their inaccuracy for species identification, particularly for subspecies differentiation. For example, biochemical tests such as: Lysine decarboxilase, L-Sorbose acid, L-Proline acid, Citrate Simmons, Dulcitole, Sucrose, Lactose, Inositole, Mucate, Esculine, D-Tartrate decomposition and nitrite to nitrate reduction11. Nevertheless, Klebsiella species are able to use several carbon resources for their metabolic pathways, which can be detected by automated methods12,13. Hence, The variability of bacterial metabolic behavior justify the design of a new diagnostic tool based on genotyping high throughput molecular techniques, being the analysis of evolutionary conserved and variable genes a useful approach to this diagnostic challenge.
The occurrence of multiple biotypes on Klebsiella genus impair the differentiation of this etiologic agents on clinical samples; in fact, bacterial behavior on Red methyl, Urea, Indole production, Citrate utilization tests has been reported as unstable assays since it depend on aerobic conditions, pH, colony differences and variability between systems used for conventional identification14.
These problems described on commercial systems as API 20E, are influenced by circumstances such as: inoculum size, inoculation time, interpretation technique, and subculture which can affect the reproducibility and as consequence results are discrepant on specie identity confirmation14.
Typing difficulties of K. pneumoniae rhinoscleromatis has been recognized not only on traditional methods based on agar and broth cultures, but also on commercial methods as API 20E and recommended panels for Klebsiella differentiation15.
In this paper, we proposed an algorithm for K. pneu-moniae ozaenae identification with Ban II enzyme, follow by restriction with Sal I or Taq I for K. pneumoniae rhinoscleromatis specific discrimination (Figure 5).
At present, database availability of rDNA 16S genes for several bacteria species provides fundamental information for comparative studies by using bioinformatic tools, which can predict accurately gene polymorphism among species correlating with phenotypic features.
Alignments allowed to recognize that rDNA 16S gene is highly conserved on Klebsiella genus, which is concordant with high similarity values (>99%) reported at intra specific level for this taxonomic group, except for K. pneumoniae and K. oxytoca. Besides this finding, phylogenetic relationship for K. pneumoniae subspecies in a single branch has been reported9.
Variability rate among rDNA 16S genes for K. pneumoniae rhinoscleromatis was found, which could be explained by the differential band patterns identified with Sal I and Taq I restriction enzymes and the reason can be attributable to the seven 16S ADNr gene copies in the genus1.
Genetic heterogeneity occurrence has been previously described on K. oxytoca associated with polymorphism on constitutive genes such as rDNA 16S, RNA polymerase subunit-b (RpoB), repetitive enterobacterial intergenic consensus sequences (ERIC-1R) and b-lactamases encoded chromosomally (OXY-1 y OXY-2), leading to recognition of two genetic subtypes into a same specie16. Moreover, technical approach to explain intraspecific variation could be not applicable to DNAr 16S sequences only for one strain not necessarily derived on taxonomic determination like subspecies or new prokaryotic species17.
Finally, recent modifications on taxonomy classification of Klebsiella genus has sketched out the needs to study the molecular epidemiology, diversity and pathogenesis for Klebsiella clinical and environmental isolates because of their differential expression of virulence factors18,19.
The proposed method for K. pneumoniae discrimination at subspecies level might generate low band patterns which help outcome interpretation.
Restriction enzymes proposed for this molecular assay are commercially available and can be economically favorable in comparison with amplified fragment length polymorphism (AFLP), pulsed field gel electrophoresis (PFGE) and randomly amplified polymorphic DNA (RAPD) which can provide low reproducibility and became difficult to interpret20; however we state out a need for validation of this technique with reference strains.
Early diagnosis of ozena and rhinoscleroma through a fast and specific molecular method with high discriminatory power at subspecies level might contribute to a rapid therapeutic formulation and thus leading to improvement of patient prognosis, avoiding destructive consequences and sequelae on the respiratory mucosa.
ACKNOWLEDGEMENTS
Author’s express special thanks to Inés Cuervo and Elizabeth Torres from Centro de Investigaciones-Biomédicas (CIBM), School of Health Sciences, Universidad del Quindío for their technical assistance on phenotypic typing of bacterial strains included in this study.
REFERENCES
1. Martínez J, Martínez L, Rosenblueth M, Silva J, Martínez-Romero E. How Are gene sequence analysis modifying bacterial taxonomy? The case of Klebsiella. Int Microbiol. 2004; 7: 261-8.
2. Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: Epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. 1998; 11: 589-603.
3. Drancourt M, Bollet C, Carta A, Rousselier P. Phylogenetic analyses of Klebsiella species delineate Klebsiella and Raoultella gen nov., with description of Raoultella ornithinolytica comb. nov., Raoultella terrigena comb. nov. and Raoultella planticola comb. nov. Int J Syst Evol Microbiol. 2001; 51: 925-32.
4. Matsen JM, Spindler JA, Blosser RO. Characterization of Klebsiella isolates from natural receiving waters and comparison with human isolates. Appl Microbiol. 1974; 28: 672-8.
5. Sahly H, Podschun R. Clinical, bacteriological, serological aspects of Klebsiella infections and their spondylarthropathic sequelae. Clin Diagn Lab Immunol. 1997; 4: 393-9.
6. Andraca R, Edson RS, Kern EB. Rhinoscleroma: A growing concern in the United States? Mayo Clinic Experience. Mayo Clin Proc. 1993; 68: 1151-7.
7. Artiles F, Bordes A, Conde A, Domínguez S, Ramos JL, Suárez S. Rinitis crónica atrófica e infección por Klebsiella ozaenae. Enferm Infecc Microbiol Clin. 2000; 18: 299-300.
8. Maguiña C, Cortés-Escalante J, Oseres-Plengue F, Centeno J, Guerra H, Montoya M, et al. Rhinoscleroma: eight Peruvian cases. Rev Inst Med Trop S Paulo. 2006; 48: 295-9.
9. Boye K, Hansen D. Sequencing of 16s rDNA of Klebsiella: taxonomic relations within the genus and to other Enterobacteriaceae. Int J Med Microbiol. 2003; 292: 495-503.
10. Stephen AF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997; 25: 3389-402.
11. Koneman EW, Allen SD, Janda WM, Schreckenberger PC, Winn WC. Diagnóstico microbiológico. Texto y atlas a color. 5ª ed. Protocolos 1258-1357. Buenos Aires: Editorial Médica Panamericana; 2001.
12. Tang YW, Ellis NM, Hopkins MK, Smith DH, Dodge DE, Persing DH. Comparison of phenotypic and genotypic techniques for identification of unusual aerobic pathogenic Gram-negative bacilli. J Clin Microbiol. 1998; 36: 3674-9.
13. Marica D, Kadar I, Rudareanu N, Kadar L, Corlatean M, Miclea I, et al. Aerobic conditions and the dynamics of the methyl red reaction in Klebsielleae. Rev Ig Bacteriol Virusol Parazitol Epidemiol Pneumoftiziol Bacteriol Virusol Parazitol Epidemiol. 1990; 35: 71-6.
14. Berlutti F, Thaller MC, Pezzi R. Unusual behaviour of Klebsiella rhinoscleromatis strains on API 20E strips. Microbiology. 1988; 11: 77-80.
15. Hansen DS, Aucken HM, Abiola T, Podschum R. Recommended test panel for diferentiation of Klebsiella species on the basis of a trilateral interlaboratory evaluation of 18 biochemical tests. J Clin Microbiol. 2004; 42: 3665-9.
16. Granier SA, Plaisance L, Leflon-Guibout V, Lagier E, Morand S, Goldstein FW, et al. Recognition of two genetic groups in the Klebsiella oxytoca taxon on the basis of chromosomal beta-lactamase and housekeeping gene sequences as well as ERIC-1 R PCR typing. Int J Syst Evol Microbiol. 2003; 53: 661-8.
17. Clayton RA, Sutton G, Hinkle PS JR, Bult C, Fields C. Intraspecific variation in small-subunit rRNA sequences in genbank: why single sequences may not adequately represent prokaryotic taxa. Int J Syst Bacteriol. 1995; 45: 595-9.
18. Drancourt M, Bollet C, Carlioz A, Martelin R, Gayral JP, Raoult D. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J Clin Microbiol. 2000; 38: 3623-30.
19. Podschun R, Pietsch S, Holler C, Ullmann U. Incidence of Klebsiella species in surface waters and their expression of virulence factors. Appl Environ Microbiol. 2001; 67: 3325-7.
20. Olive DM, Bean P. Principles and applications of methods for DNA-based typing of microbial organisms. J Clin Microbiol. 1999; 37: 1661-9.