Human papillomavirus (HPV) detected in restored plasma DNA from women diagnosed with pre-invasive lesions and invasive cervical cancer
Yazmín Rocío Arias, MSc1,2, Edward Fabián Carrillo, PhD1, Fabio Ancízar Aristizábal, PhD1
1. Department of Pharmacy, Universidad Nacional de Colombia, Bogotá, DC, Colombia.
e-mail: faaristizabalg@unal.edu.co efcarrillob@unal.edu.co yarias@colsanitas.com
2. Laboratory of Molecular Biology, Clínica Colsanitas, Bogotá, DC, Colombia.
Received for publication December 7, 2009 Accepted for publication January 12th, 2010
SUMMARY
Objective: To improve the sensitivity of Human Papillomavirus (HPV) detection in plasma from high-grade cervical neoplasia patients (CIN III) and cervical cancer (CC) evaluating any likely correlation with disease stage.
Method: We subjected plasma DNA isolates from 112 patients (CIN and ICC) to a pre-PCR restoration treatment to improve detection sensitivity. HPV-specific sequences were detected by conventional PCR both in cervical scrapes and plasma DNA obtained from each patient. For every single DNA sample, both non-restored and restored isolates were PCR analyzed.
Results: We detected HPV in plasma DNA isolates with significantly higher efficiency on restored plasma-DNA as compared to each non-restored equivalent, still maintaining close correlation with the clinical stage of the cases. By analyzing plasma-DNA isolates we could classify as HPV positive >50.0% of the cases that were previously known to be positive from the cervical scrape based assay. Interestingly, 100% of the cases in which subtype HPV18 was detected in cervical scrapes were also positive in plasma DNA.
Conclusions: Restoration of plasma DNA from cervical cancer patients allows a more sensitive PCR-based HPV detection, maintaining the correlation to disease stage traditionally observed.
Keywords: Cervical cancer; HPV; Cervical intraepithelial neoplasia (CIN); Plasma DNA.
Detección del virus del papiloma humano (VPH) en ADN de plasma restaurado de mujeres en quienes se diagnosticaron lesiones pre-invasivas y cáncer cervical invasivo
RESUMEN
Objetivo: Mejorar la sensibilidad y la detección del virus del papiloma humano (VPH) en el plasma de pacientes con neoplasias intraepiteliales de alto grado (NIEAG) y cáncer de cuello uterino (CCU) para evaluar si existe una relación con el estadío de la enfermedad.
Método: Los ADN de plasma aislados de 112 pacientes (NIC y CCU) se sometieron a restauración mediante reacción de la polimerasa en cadena (siglas en inglés, PCR), para mejorar su calidad como sustrato para PCR. En cada paciente se detectaron secuencias específicas del VPH por PCR convencional, tanto en exudados cérvico-vaginales como en ADN del plasma. Por cada muestra se analizaron por PCR, cantidades equivalentes del ADN aislado, tanto no restaurado como restaurado.
Resultados: Para las muestras pareadas se pudo detectar VPH en ADN de plasma de forma más eficiente en los materiales restaurados, pues se mantuvo una estrecha correlación con el estadío clínico de los casos. Mediante el análisis de ADN de plasma es posible detectar como VPH positivas más de 50% de los casos que se identificaron previamente como positivos en citología de cuello uterino. Se enfatiza el hecho que 100% de los casos en los que el subtipo VPH18 fue descubierto en exudado cérvico-vaginal también fueron positivos en el ADN de plasma.
Conclusiones: El proceso de restauración de ADN de plasma de pacientes con cáncer de cuello uterino permite mejorar la detección de VPH, por PCR, y mantener la correlación con el estadío de la enfermedad.
Palabras claves: Cáncer de cuello uterino; VPH; Neoplasia intra-cervical (NIC); ADN de plasma.
Cervical cancer (CC) is the second most prevalent cancer among females around the world1. CC is the major oncological problem in Colombia with an annual incidence of 36.4 new cases per 100,000 females (IARC, 2002). More than 80% of the patients have advanced tumors when they are diagnosed, whilst only 20% of cases are diagnosed at stage I2,3. Human papillomavirus (HPV), the main associated pathogenic agent, is present in 90%-100% of the cases and is believed to be a necessary, although not sufficient cause for the development of invasive cervical carcinomas4. On the other hand, previous studies have established that human blood plasma from cancer patients contains increased amounts of nucleic acids whose origin and release mechanism are not yet clear5,6.
Reports concerning HPV detection in plasma or sera from CC patients have yielded varying results so that reported plasma DNA detected concentrations span a wide range of values7-9. Bonin et al., demonstrated that DNA quality from archival clinical samples can be significantly improved by a pre-PCR restoration treatment, providing better amplification efficiencies10. In a previous work, we found this strategy to be useful for the detection of a human single-copy gene in DNA isolates from archived plasma samples11. Since plasma from cervical cancer patients contains detectable quantities of HPV-specific DNA9,12,13, we decided to evaluate the effect of applying the restoration treatment to plasma DNA isolates from CIN III and CC patients on the HPV detection frequency. HPV is detected at a very high frequency by PCR (close to 100%) in these kinds of patients taking cervical scrapes as the source of template DNA. However, most of articles focused on detection in plasma DNA show a lower and variable detection rate14,15, suggesting that different sample processing, preservation and/or different detection technologies can influence the results.
MATERIALS AND METHODS
Study population. In this study, 112 cases diagnosed at two third-level hospitals in Bogotá, Colombia, were included. None of them had started any type of treatment before being asked to participate in the study. Samples were collected during 2004 and 2005. The whole study and the consent form were prepared according to the Colombian Ministry of Health Resolution 008430, and approved by the ethics committees of both participant health institutions and the research committee of the Faculty of Science at Universidad Nacional de Colombia.
Patient age ranged from 22 to 84 years (average age=50 years). Most patients were between 41 and 50 years of age (32.1%); while the group between 51 and 60 years of age included 24.1% of the patients. 43.8% of these women had only one sex partner, while 28.6% declared 2-3 and 18.8% reported more than 4 partners. Among our group we observed an average of 5 pregnancies.
Their histopathological diagnosis revealed 25 cases (22.3%) as CIN III, 85 cases (75.9%) as squamous cell cancer stages I to IV and 2 cases (1.8%) diagnosed as adenocarcinoma stage IIIB. Twenty healthy controls were chosen among women with negative cytological results at the time of sampling.
Sample collection and DNA isolation. Every patient and healthy control provided two samples: a cervical scrape and a blood sample. Cervical scrapes and Peripheral blood samples were collected and immediately processed as previously described2,11. Both cervical scrape and plasma samples were extracted by using a QIAamp DNA Mini Kit (QIAGEN; Valencia, CA, USA) following the manufacturer’s instructions. However, for plasma samples the protocol was modified by adding 1 ml glycogen (SIGMA, Milwaukee, WI, USA) along with the Ethanol aliquot indicated at the step just before column loading. DNA was finally eluted with 100 ml of AE buffer.
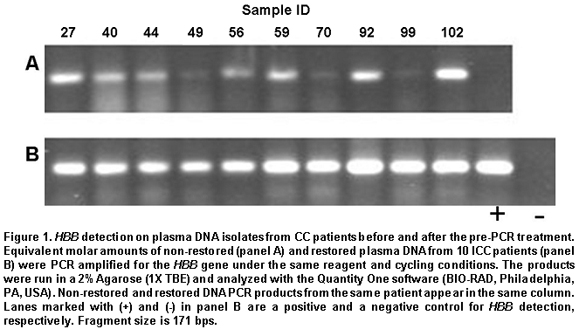
Pre-PCR restoration treatment. Before PCRs were carried out, each DNA isolate was subjected to a pre-PCR restoration treatment, as previously reported11, to improve PCR sensitivity when detecting HPV specific sequence. Detection results obtained by PCR on restored and non-restored plasma DNA isolates from 10 patients were compared to determine the convenience of this treatment. Genes amplified at this point were the human b-globin gene (HBB) and the viral L1 gene, which provided a tool for the generic (non-subtype specific) detection of HPV.
Evaluating DNA quality. The integrity of DNA from cervical scrape and plasma samples was evaluated by amplification of a 171-bp fragment from the HBB gene as DNA quality indicator. Primer sequences for DNA quality assessment were as follows:
Glo1: 5’-AGCAACCTCACAAACAGACACC-3’
Glo2: 5’-CTACACATGCCCAGTTTC-3’
PCR reactions were carried out in a final volume of 25 ml with the following reagent concentrations: 10 mM Tris-HCl (pH 9.0 at 25°C), 50 mM KCl and 0.1% Triton® X-100, 3 mM MgCl2, 0.2 mM each dNTP, 0.3 mM of both direct and reverse primers, and 1 U of Taq DNA polymerase. All reagents were purchased from the same manufacturer (PROMEGA, Madison, WI, USA).
The reactions were carried out in a My Cycler thermal cycler (BIORAD, Hercules CA, USA) with the following protocol: 3 min at 94 ºC, followed by 35 amplification cycles of 94 ºC for 20 sec, 58 ºC for 20 sec and 72 ºC for 20 sec, and a final elongation step at 72 ºC for 5 min. Peripheral Blood Lymphocytes DNA was used as positive control.
PCR for HPV. Generic HPV was assayed only in DNA isolates from cervical scrapes and restored plasma DNAs returning positive results for the HBB gene. Generic HPV detection was accomplished by PCR using GP5+/GP6+ primers, following a procedure already described16.
After generic HPV detection, subtypes 16 and 18 were specifically detected with primers targeted for the HPV E6/E7 region previously described14. Each primer set allowed clear detection of its target sequence when using 100 picograms of control plasmid (pBR322/HPV16 or pBR322/HPV18). The length of the segments amplified were 100 bp for HPV16 and 126 bp for HPV18.
RESULTS
Plasma DNA restoration and PCR detection. We evaluated DNA quality improvement by trying the procedure on plasma DNA from 10 CC cases and comparing amplification results for HBB on treated and non-treated DNA. Amplification efficiency was notoriously increased by the treatment, as judged by observed band intensities (Figure 1). Density analysis in Figure 2A shows that band intensity measurements after restoration have lesser inter-sample variability than values without treatment. This change is observed because the general tendency is that samples with low initial detection levels showed greater increases upon restoration than those with initial high values. Similar results were observed regarding HPV generic detection, as shown in Figure 2A. Detection sensitivity was also improved when using GP5+/GP6+ primers for the L1 region in the viral genome. Interestingly, in this group of samples, only two were detected as HPV positive when non-restored DNA was assayed (samples 44 and 92), the assay performed on the corresponding restored isolates confirmed theses results and allowed viral detection in 2 more samples (samples 49 and 99). So, the restoration treatment allowed the classification as positive of two samples considered negative at the first assay.
Detecting HPV in cervical scrapes and plasma samples. DNA quality in both plasma and cervical scrape isolates was evaluated by amplification of a single-copy gene (HBB). Good-quality DNA from both plasma and cervical scrape was obtained only from 107 out of 112 cases, 5 were discarded. HPV was assayed in 107 cervical scrapes. Prevalence found was 95.3% (102/107). Among these samples there were 25 cases diagnosed as CIN III. When cervical scrapes from them were assayed for HPV, 23 could be classified positive (92%). On the other hand, HPV could be detected in 48.6% of the corresponding plasma DNA isolates (52/107). As expected, viral detection in plasma DNA is less efficient than in cervical scrapes.
If only cases positively detected in cervical scrapes are considered (102 cases), the subgroup of cases also found positive for HPV in plasma DNA (restored DNA) is 51%. Next, we assayed our samples specifically for HPV 16 and 18 in scrapes and plasma. When the 102 HPV-positive cervical scrapes were assayed, 55 of them resulted HPV-16 positive (54%). HPV-18 prevalence was only 8.8% among our cases (9 cases), a result closely resembling findings by Molano et al.3. Also, viral types detected in plasmas always resembled the types previously found in matched cervical scrapes. In no case we detected any viral subtype in plasma not previously demonstrated in the corresponding cervical scrape. HPV 18 is the predominant HPV type in adenocarcinomas17. Our pool of samples included only two invasive adenocarcinoma cases, both of which were HPV-18 positive. After restoration, HPV 16 was detected in plasma in 30 of the 52 HPV-positive cases (57.7%). HPV18 was detected in plasma with the same frequency it was detected in cervical scrapes (100%). None of the 20 control cases, healthy women with negative cytology, yielded positive HPV detection when their scrape and plasma DNA isolates were assessed.
The clinical stage of the disease and HPV detection in plasma DNA. Generic HPV was detected in plasma DNA in 51% of ICC cases, in 2 available adenocarcinoma cases, and in 27.3% of patients suffering from high-degree CIN III. Table 1 shows the prevalence of HPV in plasma DNA isolates from all the cases assayed according to the clinical stage. HPV prevalence was proportionally higher in those cases having greater disease grade as confirmed by the c2 test.
DISCUSSION
HPV is one of the main factors associated to cervical cancer development4; however, variable detection frequencies have been reported in cell scrapes or cervical biopsy-based studies, generally between 75 and 100%18-20. We found a very high HPV prevalence among our cases since generic detection, based on the detection by PCR of the L1 region of the viral genome in cervical scrapes, resulted positive in 95.3% of the cases (102 out of 107 cases). Our results regarding both for generic (all subtypes) and specific (subtypes 16 and 18) detection in cervical scrapes are similar to previous studies by Molano et al.2,3. In addition, those studies reported high-risk HPV 56, 58 and low-risk 81 as the most prevalent types performed also in Colombian women with negative cytology, all of them at frequencies higher than HPV 16. Here we found that HPV 16 is present at a significant frequency when abnormal growth occurs in CIN III and CC patients.
Based on our previously published results concerning the convenience of plasma DNA restoration to improve its quality as PCR template11, we decided to apply this procedure to detect HPV-specific sequences on plasma DNA isolates from cervical cancer patients. The procedure proved to notably increase HPV detection sensitivity in four out of ten test samples, two of which had been classified as negative when the restoration treatment had not been included. Based on these results, we decided to apply the restoration treatment to the 112 plasma DNA isolates included in the study. In most of the previous reports where HPV is detected by conventional PCR in matched plasma DNA and tumor DNA isolates, the detection rate in plasma ranges between 6.9%-20% of all HPV positive cases9,12. The procedure presented here provides an increased detection rate of >50%. However, we mention a report by Dueñas et al.21 and another by Wei et al.22, reporting that the virus was detected in plasma samples in 70% and 65%, respectively, of the cases where tumors were confirmed positive. The second study refers to a small number of patients (17 patients). In order to maintain costs as low as possible for a possible HPV-detection clinical assay, this study was limited to a conventional PCR analysis; nevertheless, it is a logical consequence that performing these analysis by real-time PCR could improve sensitivity and detection rate in restored plasma DNA. In no case did we detect HPV in plasma DNA not having detected it in the corresponding cervical scrape.
As stated above, 52 cases were HPV positive when the generic test was performed on plasma DNA isolates. HPV 16 was specifically detected in 30 of these cases (57.7%). Considering our global results, HPV-16 detection on restored plasma DNA was approximately half as efficient as the detection in scrapes. Interestingly, HPV18 was detected in plasma with the same frequency observed in cervical scrapes. Nine cases previously found to be positive, based on cervical scrapes were also shown as positive when assayed in plasma DNA. When analyzing plasma DNA isolates from patients presenting metastasis (15 cases), we found 12 of them positive for generic HPV (80%) and 8 of them positive for HPV 16. Three remaining cases were HPV negative at the time of the study.
Finally, we were interested in the frequency of mixed HPV 16/18 infections among our samples. Simultaneous presence of HPV 16 and 18 types was identified only in 4 cases when cervical scrape DNA was assayed. However, mixed infection could be demonstrated only in 2 of them when corresponding plasma DNA isolates was analyzed. Interestingly, HPV 18 was the type detected in both scrape and plasma from these four cases; while in two cases, HPV 16 could only be detected in the scrape. This fact could suggest different release rates of viral DNA to the plasma between types. One could hypothesize that viral DNA integration into the host genome is a condition promoting its detection by PCR in plasma DNA. It would be released along with the host genomic DNA, being subjected to the same degradation processes. But this idea must be tested.
HPV detection in restored plasma DNA clearly increased as the disease’s progression level increased; 27.3% positivity was found in high-grade pre-neoplastic lesions (CIN III); whilst it reached 90% in patients suffering from stage IV ICC. Regarding early stages, one could hypothesize that HPV is actually present in the remaining CIN III, HPV-negative cases although at doses below the detection limit. Even though the real time PCR is a good alternative, implementation of new, more sensitive and low cost detection techniques will be an important objective if HPV detection in blood is to be applied on very early neoplastic stages (CIN I and II) or even screening programs on healthy women.
Conflict of interest. None of the authors has conflicts of interest related to this study.
ACKNOWLEDGEMENTS
We thank the Department of Pharmacy and the Institute of Biotechnology at Universidad Nacional de Colombia (Bogotá, Colombia) for providing their facilities for this research. This study was supported by the Division of Research at Universidad Nacional de Colombia (Resolution 347, 22 February 2006).
REFERENCES
1. Longworth MS, Laimins LA. Pathogenesis of human papillomaviruses in differentiating epithelia. Microbiol Mol Biol Rev. 2004; 68: 362-72.
2. Molano M, Posso H, Weiderpass E, van den Brule AJ, Ronderos M, Franceschi S et al. Prevalence and determinants of HPV infection among Colombian women with normal cytology. Br J Cancer. 2002; 87: 324-33.
3. Molano M, van den Brule AJ, Posso H, Weiderpass E, Ronderos M, Franceschi S et al. Low grade squamous intra-epithelial lesions and human papillomavirus infection in Colombian women. Br J Cancer. 2002; 87: 1417-21.
4. Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999; 189: 12-9.
5. Sidransky D. Circulating DNA. What we know and what we need to learn. Ann NY Acad Sci. 2000; 906: 1-4.
6. Stroun M, Maurice P, Vasioukhin V, Lyautey J, Lederrey C, Lefort F et al. The origin and mechanism of circulating DNA. Ann NY Acad Sci. 2000; 906: 161-8.
7. Widschwendter A, Blassnig A, Wiedemair A, Muller-Holzner E, Muller HM, Marth C. Human papillomavirus DNA in sera of cervical cancer patients as tumor marker. Cancer Lett. 2003; 202: 231-9.
8. Yang HJ, Liu VW, Tsang PC, Yip AM, Tam KF, Wong LC et al. Quantification of human papillomavirus DNA in the plasma of patients with cervical cancer. Int J Gynecol Cancer. 2004; 14: 903-10.
9. Dong SM, Pai SI, Rha SH, Hildesheim A, Kurman RJ, Schwartz PE et al. Detection and quantitation of human papillomavirus DNA in the plasma of patients with cervical carcinoma. Ca Epidemiol Biomarkers Prev. 2002; 11: 3-6.
10. Bonin S, Petrera F, Niccolini B, Stanta G. PCR analysis in archival postmortem tissues. Mol Pathol. 2003; 56: 184-6.
11. Arias YR, Carrillo EF, Aristizábal FA. Plasma DNA restoration for PCR applications. J Clin Pathol. 2007; 60: 952-4.
12. Capone RB, Pai SI, Koch WM, Gillison ML, Danish HN, Westra WH et al. Detection and quantitation of human papillomavirus (HPV) DNA in the sera of patients with HPV-associated head and neck squamous cell carcinoma. Clin Cancer Res. 2000; 6: 4171-5.
13. Pornthanakasem W, Shotelersuk K, Termrungruanglert W, Voravud N, Niruthisard S, Mutirangura A. Human papillomavirus DNA in plasma of patients with cervical cancer. BMC Ca. 2001; 1: 2.
14. Liu VW, Tsang P, Yip A, Ng TY, Wong LC, Ngan HY. Low incidence of HPV DNA in sera of pretreatment cervical cancer patients. Gynecol Oncol. 2001; 82: 269-72.
15. Sathish N, Abraham P, Peedicayil A, Sridharan G, John S, Shaji RV et al. HPV DNA in plasma of patients with cervical carcinoma. J Clin Virol. 2004; 31: 204-9.
16. de Roda Husman AM, Walboomers JM, van den Brule AJ, Meijer CJ, Snijders PJ. The use of general primers GP5 and GP6 elongated at their 3' ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. J Gen Virol. 1995; 76: 1057-62.
17. Leminen A, Paavonen J, Vesterinen E, Wahlstrom T, Rantala I, Lehtinen M. Human papillomavirus types 16 and 18 in adenocarcinoma of the uterine cervix. Am J Clin Pathol. 1991; 95: 647-52.
18. Muñoz N. Human papillomavirus and cancer: the epidemiological evidence. J Clin Virol. 2000; 19: 1-5.
19. zur Hausen H. Molecular pathogenesis of cancer of the cervix and its causation by specific human papillomavirus types. Curr Top Microbiol Immunol. 1994; 186: 131-56.
20. Cuschieri KS, Cubie HA. The role of human papillomavirus testing in cervical screening. J Clin Virol. 2005; 32 (Suppl 1): 34-42.
21. Dueñas A, Lizano M, Carrillo AL, Wójcik J, Trejo C. DNA of human papillomavirus detected by PCR in plasma of cervical cancer patients: a potential marker of residual disease. Clin Chem. 2001; 47: 364.
22. Wei YC, Chou YS, Chu TY. Detection and typing of minimal human papillomavirus DNA in plasma. Int J Gynaecol Obstet. 2007; 96: 112-6.