Analysis of HLA-A, HLA-B, HLA-DRB1 allelic, genotypic, and haplotypic frequencies in colombian population
Yazmin Rocío Arias-Murillo, Bacteriol, MSc1,2, Miguel Ángel Castro-Jiménez, MD, MSc2,3, María Fernanda Ríos-Espinosa, Bacteriol1, Juan Javier López-Rivera, MD, MSc1, Sandra Johanna Echeverry-Coral, Bacteriol Esp1, Oscar Martínez-Nieto, MD4
1. Laboratory of Molecular Biology, Clinical Pathology Research Group (INPAC), Clínica Colsanitas SA, Bogotá, DC, Colombia. e-mail: ariasmurillo@hotmail.com mafecita74@hotmail.com jlopez@colsanitas.com
jecheverry@colsanitas.com
2. Grupo Colombiano de Estudios Alfa en Epidemiología, Salud Poblacional, Estadística Aplicada y Ciencias Aliadas, Bogotá, DC, Colombia. e-mail: mcastro2505@yahoo.es
3. Grupo de Virología, Red Nacional de Laboratorios, Instituto Nacional de Salud, Bogotá, DC, Colombia.
4. Clinical Laboratory Medical Director, Clinical Pathology Research Group (INPAC), Clínica Colsanitas SA, Bogotá, DC, Colombia. e-mail: osmartin@colsanitas.co
Received for publication September 19, 2009 Accepted for publication May 25, 2010
SUMMARY
Introduction: The high polymorphism of the HLA system allows its typification to be used as valuable tool in establishing association to various illnesses, immune and genetic profiles; it also provides a guide to identifying compatibility among donors and receptors of organs transplants.
Objective: To establish HLA-A, HLA-B, and HLA.DRB1 allele, genotype and haplotype frequencies among patients treated at Clinica Colsanitas SA.
Methods: 561 patients coming from different regions in Colombia, who were attended in 8 centers of the clinical laboratory of the Clinica Colsanitas in different cities of the country from January 2004 to August 2008, were included in this study. All were HLA-A,-B, and -DRB1 typified via SSP PCR. Allele, genotype and haplotype frequencies were estimated with STATA Software Version 9.0 and the GENEPOP genetic analysis package.
Results: 19, 28, and 15 different alleles were identified for loci HLA-A,-B and -DRB1, respectively. Alleles found most frequently were A*24 (26.2%), A*02 (26%), B*35(22.7%), and DRB1*04 (24%). The most frequent genotypes were A*02,24 (14.2%), B*07,35 (5.5%), DRB1*01,04, and DRB1*04,04 (6.9%); while most the frequent haplotypes were HLA A*24, B*35 (9.2%), A*24, DRB1*04 (8.1%); B*35, DRB1*04 (7.8%), A*2 DRB1*04 (7.4%).
Conclusion: The results obtained provide a useful reference framework for the population studied, allowing compatibility probability calculations to be performed for organ transplants.
Keywords: HLA; Genotype; Haplotype; Allele; Gene.
Análisis de frecuencias alélicas, genotípicas y haplotípicas HLA -A, HLA-B, HLA-DRB1 en población colombiana
RESUMEN
Introducción: El alto polimorfismo del sistema HLA, hace que su tipificación sea una herramienta de gran valor al establecer asociación con diferentes enfermedades, patrones inmunológicos, antropogenéticos, así como para establecer probabilidades de encontrar donantes compatibles con receptores de diferentes tipos de trasplante de órganos.
Objetivo: Establecer las frecuencias alélicas, genotípicas y haplotípicas en pacientes atendidos en la Clínica Colsanitas SA.
Metodología: Se incluyeron un total de 561 pacientes atendidos en el Laboratorio Clínico de La Clínica Colsanitas SA, en 8 sedes en diferentes ciudades del Colombia, durante el período comprendido entre enero de 2004 a agosto de 2008. Se realizó tipificación de HLA -A,-B,-DRB1 por PCR SSP. Las frecuencias alélicas, genotípicas y haplotípicas fueron estimadas mediante el paquete estadístico Stata y el paquete de análisis genético Genepop.
Resultados: Fue posible la identificación de 19, 28 y 15 alelos de los loci HLA A-B-DRB1 respectivamente, de los cuales los más frecuentes fueron A*24 (26.2%), A*02 (26%), B*35 (22.7%), DRB1*04 (24%). Los genotipos más frecuentes encontrados fueron A*02,24 (14.2%), B*07,35 (5.5%), DRB1*01,04 y DRB1*04,04 (6.9%). Los haplotipos más frecuentes fueron: HLA A*24, B*35 (9.2%), A*24, DRB1*04 (8.1%); B* 35, DRB1*04 (7.8%), A*2 DRB1*04 (7.4%).
Conclusión: Los resultados obtenidos permiten tener referencia para aplicaciones en la población estudiada, así como para establecer probabilidades de compatibilidad en la creciente área de trasplante de órganos.
Palabras clave: HLA; Genotipos; Haplotipos; Genes; Alelos.
The human leukocyte antigen system (HLA) of the major histocompatibility complex (MHC) contains a numerous family of genes located on the short arm of the 6th chromosome (6p21.1-6p21.3). These antigens are expressed in autosomal and codominant manner and they are inherited from both parents, which is why each individual expresses an antigen of paternal origin and one of maternal origin for each of the loci1.
The high polymorphism of the HLA system and advancements in molecular biology have permitted for the typifying of these antigens to become important tools in establishing associations of susceptibility or resistance to disease, development of tumor processes; playing an important role in understanding the variability of the immune response and determinant on the survival of organ or tissue transplants2-8.
Because the allele frequencies and the disequilibrium patterns vary within a given population, the characterization of these molecules and the determination of their frequencies gain much pertinence and permits establishing parameters of association to disease in the population, which has a predictive value in managing self-immune diseases like Systemic Erythematosus Lupus (SEL) and Rheumatoid Arthritis (RA), also becoming an important tool for the distribution of organs donated in the different transplant groups in the National Donation Network, as well as having an additional usefulness in estimating times the receptors must remain in the waiting list9,10.
Most of the Latin American populations studied are of indigenous origin or barely mixed11-15. The population treated in the Molecular Biology laboratory at Clinica Colsanitas SA for HLA typifying comes from different Colombian cities, offering an opportunity to determine the allele, genotype, and haplotype frequencies in patients of diverse origins.
MATERIALS AND METHODS
Design and population. This is a retrospective descriptive study, which included 561 patients treated in the cities of Bogota, Cali, Barranquilla, Medellin, Villavicencio, Armenia, Bucaramanga, and Neiva. The patients underwent HLA A,-B and -DRB1 typifying in the Molecular Biology laboratory at Clinica Colsanitas between January 2004 and August 2008.
Definition of variables. The variables of interest for this study were: demographic characteristics (gender, age, place of birth), alleles for each of the 3 loci analyzed (HLA A*, HLA B*, HLA DRB1*), as well as the genotypes and haplotypes found.
Procedure of HLA typifying. DNA extraction was carried out with the Wizard® Genomic DNA Purification kit, adhering to the protocol recommended by the manufacturer from the white blood cell layer obtained from blood samples anticoagulated with EDTA or from spleen fragments contained in sterile containers, hermetically sealed in an RPMI medium supplemented with fetal calf serum.
The final concentration for each of the samples was adjusted at 120 ng/µl and then the HLA-A, HLA-B, and HLA-DRB1 genes were determined via single specific primer polymerase chain reaction (SSP PCR) as per manufacturer instructions (Biotest ABDR SSPtray). Thereafter, electrophoresis was conducted on the amplification products in agarose gels at 2% dyed with SYBR® Safe DNA gel (Invitrogen). An analysis was performed of amplified bands by using Quantity One (BIORAD), leaving a photo record and the allele assignation was made by employing the tables provided, as well as the Biotest SSP typing software.
Statistical analysis. Proportions were calculated to summarize the qualitative variables. The normality of the age variable was evaluated by using the Shapiro-Wilk test and summarized with a mean.
Homologation of the degree of resolution of the typifying to two digits was done on the data base for further analysis of allele, genotype, and haplotype frequencies16.
The p values lower than or equal to 0.05 were considered significant. The allele, genotype, and haplotype frequencies and those corresponding to demographic characteristics were calculated in the Stata 9.0/SE program. The Linkage disequilibrium between loci pairs, the Fisher fixation index, and the Hardy Weinberg equilibrium were estimated by employing the GENEPOP genotype analysis package version 1.217, which bears in mind the modifications to the Markov chain algorithm described by Guo and Thompson17.
Ethical considerations. This study used data from patients whose samples had already been taken and processed; hence, there was no additional risk. The confidentiality of the information was strictly protected. The samples had been taken after obtaining signed informed consent from the patients or their families and through medical request.
RESULTS
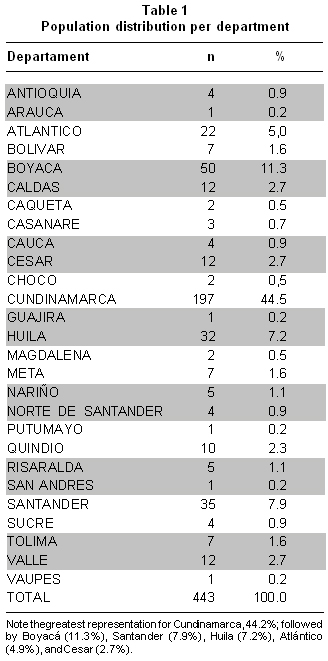
Demographic characteristics. Of the total number of patients included in the study, we obtained information on the place of birth from 443 patients; who came from 28 departments in Colombia, with a greater representation from Cundinamarca, followed by Boyaca, Santander, Huila, Atlantico, and Cesar. The population distribution per department can be seen in Table 1.
Of the individuals included, 57.4% were male and 42.6% female, the mean age was 38 years of age with a range from 1 to 85 years. Within the population analyzed, there were patients who attended outpatient consultation to have the sample taken, as well as patients who were under pre-transplant study by the group at Clinica Colsanitas SA and cadaveric donors from the 1st regional facilities of the transplant network typified by the renal transplant group at Clinica Reina Sofia.
Allele frequencies. It was possible to determine 19, 28, and 15 alleles of the HLA A-B-DRB1 loci, respectively, in the population studied. The most frequent alleles in the HLA-A locus were A*24 (26.2%), A*02 (26%), A*01 (7.6%), A*68 (6.5%), and A*29 (6.4%); followed by A*03, A*39, A*33, A*11, A31, A*23, A*26, and A*32 with frequencies over 1% (Table 2).
The most frequent alleles in the HLA- B locus were B*35(22.7%), B*44 (11.1%), B*07 (8.6%), B*51(8%); followed by B*40, B*14, B*39, B*15, B*18, B*38, B*13, B*48, B*27, B49, B*58, B*57, and B*45 with frequencies over 1% (Table 3). The most frequent alleles in the HLA-DRB1 locus were DRB1*04 (24.1%), DRB1*15 (11.3%), DRB1*07 (11.1%), DRB1*01 (10.3%), and DRB1*13 (8.3%); followed by DRB1*11, DRB1*08, DRB1*14, DRB1*16, and DRB1*17 with frequencies over 3% (Table 4).
Genotype frequencies. We observed 90 different genotypes for the HLA-A locus, 144 genotypes for HLA-B, and 99 for HLA-DRB1; the most frequent genotypes for the HLA-A locus were: A*02,24 (14.2%), A*24,24 (7.3%), A*02,02(6.1%), A*02,68 (4.3%), and A*01,02 (3.95%). Graphic 1 shows the genotypes for each of the loci analyzed, which reveal frequencies over 1%.
The genotypes revealing the highest frequencies for the HLA B locus were: B*07,35 (5.53%), B*35,44 (4.63%), B*35,35 (4.28%), and B*14,35 (3.21%); for the HLA DRB1 locus, the most observed genotypes were: DRB1*01,04 and DRB1*04,04 (6.95%); followed by DRB1*04,15 (4.3%), DRB1*04,13(4.1%), DRB1*04,11 (3.6%), and DRB1*04,07 (3.6%).
The Hardy-Weinberg equilibrium for each locus was tested through the GENEPOP program, obtaining a deficiency of heterozygous individuals for DRB1 genotypes with p: 0.0091 and reaching a Fisher fixation index of + 0.0344; while the genotype frequencies for the HLA A and HLA B loci were within the Hardy-Weinberg equilibrium (Table 5).
The linkage disequilibrium values among the haplotypes analyzed were significant, obtaining p= 0.00992 between the HLA A and HLA B loci, p= 0.0012 for the HLA A and HLA DR loci, and p= 0.026 for the HLA B and HLA DR loci.
Haplotype frequencies. The most commonly found HLA A, B haplotypes in the population studied were: HLA A*24 B*35 (9.2%); followed by A*02 B35 (5.2%), A*02 B*51 (3.1%), A*24 B*40 (2.9%), A*02 B*44 (2.68%). The most frequent HLA B, DR haplotypes were: HLA B*35 DRB1*04 (7.8%), B*44 DRB1*07 (3.38%), B*07, and DRB1*15 (2.6%), and for HLA A, DR: A*24 DRB1*04 (8.1%), A*2 DRB1*04 (7.4%), and A*02 DRB1*15 (3.2%). The haplotypes showing frequencies over 1% are described in Graphic 2.
DISCUSSION
It was possible to determine the level of the A locus for 19 alleles, similar to those reported by Ossa et al.14, for a population from the city of Bogotá. When comparing with a study reported in cadaveric donors conducted in the city of Medellín, it was determined that the A43 and A80 alleles were not identified, and which were observed in this study. The frequencies obtained were similar for the first two places of the A* locus as those reported by Ossa et al.14, and Rodriguez et al.9, for a Colombian population; however, differences in alleles were found, like: A*01, A*68, A*29, which show higher frequencies (7.6%, 6.5%, 6.4%) with respect to that reported in the Medellin, Colombia study (5.1%, 5.6%, 3.7%).
The analyses made by Ossa et al.14, reveal similar frequencies for the HLA A locus to those reported in the current study. The differences observed in relation to the Medellín study may arise because even though it is a Colombian population, they present information from a specific city; while in our case, the individuals come from different parts of the nation, which makes it a heterogeneous population.
The results obtained in this study correlate to those previously reported for a Latin American population, where the alleles with the greatest frequency were A02, A24, A01, and A6814,15,18.
Only 28 alleles were identified for the B locus; while in prior studies conducted in Colombian populations, 43 alleles were reported for this locus9,14. This marked difference is attributable to the resolution of the technique used in typifying HLA in our study, given that is some instances such resolution did not permit differentiation of the B15, B14, and B40 splits in the allelic variants currently considered. Of the total population typified as B14 (6.2%), 3.2% were differentiated as B64, B65, and B14, where B65 corresponded to the greatest frequency, but since it was not possible to define such in a broad percentage, they were reported for the current study as B14.
The HLA B*15 corresponded to 3.3% of the population. For this allele, we were not able to identify in all typifying events the specificity B62, B63, B70, B71, B72, and B75, because of the resolution of the kit used in typifying. Thus, the B15 frequency we found is higher than that reported in previous studies in Colombia. As well as for the B14 and B15 alleles, for the B40 allele we were not able to differentiate between the B61 and B60 alleles; hence, the frequency found for B40 is a reflection of the 3 alleles (B40, B60, and B61). It is important to stress that the B*46, B*47, B*67, and B*78 alleles whose frequency is equal to or lower than 0.2 %, were not found in the population analyzed in this study, contrasting with that reported by Rodriguez et al.9, which is possible bearing in mind that the size of our sample is smaller than that analyzed in the city of Medellín.
The most common alleles for the HLA B, B*35, and B*44 loci basically correspond to that reported in prior studies in a Colombian population9 and correlate with the frequencies mostly observed in populations of Uruguay, Peru, and Argentina11,12,15; nevertheless, there are some differences with that reported by Ossa et al.14 in population from the city of Bogotá, which may be due to the heterogeneity of the population in the current study.
At the DRB1* locus level, we found 15 alleles, with the most common being: DRB1*04, DRB1*15, DRB1*07, DRB1*01, DRB1*13, DRB1*11, DRB1*08, DRB1*14, DRB1*16, and DRB1*17, which are also the ones found with the greatest frequency in prior studies in Colombian and Latin populations, although Rodriguez et al.9, found a higher frequency (13.4%) for the DRB1*13 allele and a lower frequency (10%) for the DRB1*15 allele in relation to those obtained in this study (8.3%) and (11.3%), respectively. When comparing our results to the study with the population in the city of Bogota published by Ossa et al.14, there are small differences regarding the order of the alleles found.
Upon reviewing the information on haplotypes found in the population studied, we have that the most frequent HLA A, B haplotypes were A*24 B*35, A*2 B*35, A*2 B*51, and A*29 B*44, which correlates with the reports found in studies conducted on Latin populations19; and haplotypes A*02 B*44 and A*02 B*07 with important frequencies in this study are reported most frequently in Caucasian populations. Regarding HLA B DRB1 haplotypes, the first two places were occupied by HLA B*35 DRB1*04 and B*44 DRB1*07, correlating with that reported by Rodríguez et al.9 and they are the haplotypes found in greater frequency in a Hispanic population19,20.
It is worth mentioning that since B40 is an important component of high-frequency haplotypes, it is necessary to employ techniques that permit differentiating the B60, B61, and B40 allelic variants, which this study could not establish; hence, the haplotypes in which the B40 is included may be actually made up of any of the corresponding variants and, thus, in prior studies A*24 B*61 occupy a greater frequency, which would be included in the A*24 B*40 haplotype reported in our study because of the resolution of the technique used.
The results of the current study correspond mostly to that previously reported for Colombia, where we found a typical component of Latin population with an important Caucasian influence; nevertheless, the differences evidence the variations in the size and origin of the population studied, as well as the important influence of the resolution of the technique used in typifying.
Given the heterogeneity of the population analyzed in the current study, it is important to keep the following aspects in mind: these were mostly patients attending outpatient consultation for whom typifying was requested for pre-transplant study in different cities of the nation, they belonged to groups of patients studied to be included in the waiting list for renal transplant or suspect of clinical association to a specific disease, and in a minor proportion they were cadaveric donors from the 1st regional facilities of the National Transplant Network.
All these factors may undoubtedly be associated to the results obtained in this study; however, it should be noted that of patients in renal pre-transplant study only 10% are related to self-immune diseases and, generally, HLA-disease association studies estimate that the differential association of different alleles in different populations is merely a reflection of the different distribution of frequencies of the HLA haplotypes or alleles in said populations. For this reason, more than having healthy individuals or patients in the population the origin does set an important relevant factor with respect to the distribution of frequencies.
Conducting this study permitted us to know the genotype, allele, and haplotype frequencies of the population analyzed and offers us useful tools to establish probabilities of finding compatible donors for patients in waiting lists for renal transplant. Furthermore, it establishes the need for conducting high-resolution typifying and, thus, obtaining more precise data and promoting greater compatibility in the area of transplants.
Conflict of interest. None of the authors has conflicts of interest related to this study.
REFERENCES
1. Alper CA, Larsen CE, Dubey DP, Awdeh ZL, Fici DA, Yunis EJ. The haplotype structure of the human major histocompatibility complex. Hum Inmunol. 2006; 67: 73-84.
2. Sung C.k, Don-Il H. Clinical features and prognosis of HLA-B27 positive and negative anterior uveitis in a Korean population. J Korean Med Sci. 2009; 24: 722-8.
3. Avila S, Ormsby C, Carlson J, Valenzuela H, Blanco J, Garrido D et al. Unique features of HLA-mediated HIV evolution in a Mexican cohort: a comparative study. Retrovirology. 2009; 6: 72.
4. Garavito G, Malagón C, Ramírez L, De la Cruz OF, Uribe O, Navarro E et al. Polimorfismo de los alelos de los antígenos de leucocitos humanos HLA-DRB1 y su asociación con la artritis reumatoidea juvenil en una muestra de niños mestizos colombianos. Biomedica. 2003; 23: 254-62.
5. Lim JB, Kim HO, Jeong SH, Ha JE, Jang S, Lee SG et al. Identification of HLA A*2402 restricted HCMV immediacy early-1 (IE-1) epitopes as targets for CD8+ HCMV- specific cytotoxic T lymphocytes. J Transl Med. 2009; 7: 72.
6. Da Silva SA, Mazini P, Rers P, Sell A, Tsuneto L, Peixoto R. HLA DR and DQ alleles in patients from the south of Brazil: markers for leprosy susceptibility and resistance. BMC Infect Dis. 2009; 9: 134-6.
7. Pérez V, Rocher A, Soler C, Flores C, Sarria C. Correlation in previously treated HIV-1 positive patients between hyper sensitivity reaction to abacavir and the presence of the HLA B*5701 allele. Farm Hosp. 2009; 33: 155-60.
8. Benjamin A, Bradley T. Pronostic assays for rejection and tolerance in organ transplantation. Transplant Immunol. 2005; 14: 193-201.
9. Rodríguez L, Giraldo M, García N, Velásquez L, Paris S, Alvarez C et al. Frecuencias alélicas, genotípicas y haplotípicas HLA-A, HLA-B, HLA-DRB1 en donantes fallecidos, Medellín, Colombia. Biomedica. 2007; 27: 537-47.
10. Subklewe M, Marquis R, Choquet S, Leblond V, Garnier JL, Hetzer R et al. Association of human leukocyte antigen haplotypes with posttransplant lymphoproliferative disease after solid organ transplantation. Transplantation. 2006; 82: 1093-100.
11. Alfaro E, Dipierri, J. E, Gutiérrez N, Vullo C. M, 2004, Gene and haplotype frecuencies of HLA system in the Norht-west Argentina. Antropo. 2004; 6: 15-23.
12. Bengochea M, Alvarez I, Hidalgo P, Cabrera A, Senatore O, Toledo R et al. HLA-A, -B, -DR en receptores de trasplante de médula ósea en Uruguay. Rev Med Uruguay. 2003; 19: 149-58.
13. Ossa H, Ramos O, Yunis E. Estudios genéticos de las comunidades indígenas del nororiente colombiano. Rev Fac Med Univ Nac Colomb. 1994; 42: 9-16.
14. Ossa H, Manrique A, Quintanilla S, Peña A. Polimorfismos del sistema HLA (loci A*, B* y DRB1*) en población colombiana. NOVA. 2007; 7: 25-30.
15. De Pablo R, Beraun Y, Nieto A, Calzada J, Rementeria M, Sanz L, López-Nevot M, Martín J. HLA class I and class II allele distribution in the Peruvian population. Tissue Antigens. 2000; 56: 507-14.
16. Holdsworth R, Hurley CK, Marsh S, Lau M, Noreen HJ, Kempenich M et al. The HLA dictionary 2008: a summary of HLA-A, -B, -C, -DRB1/3/4/5, and DQB1 alleles and their association with serologically defined HLA-A, -B, -C,-DR, and -DQ antigenes. Tissue Antigens. 2008; 73: 95-170.
17. Raymond M, Rousset F. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered. 1995; 86: 248-9.
18. Cao K, Hollenbach J, Shi X, Shi W, Chopek M, Fernández M. Analysis of the frequencies of HLA-A, B, and C alleles and haplotypes in the five major ethnic groups of the United States reveals high levels of diversity in these loci and contrasting distribution patterns in these populations. Hum Immunol. 2000; 62: 1009-30.
19. Mori M, Beatty PG, Graves M, Boucher KM, Milford EL. HLA gene and haplotype frequencies in the North American population: The National Marrow Donor Program Donor Registry 1. Transplantation. 1997; 64: 1017-27.
20. Comas D, Maten E, Calafell F, Pérez A, Martínez R, Bosch E, Bertranpetit J. Estudio de la variabilidad de los genes del sistema HLA en dos poblaciones de la Península Ibérica. 15ª ed. Barcelona: Secretariado de Publicaciones, Universidad de León; 2000. p. 403-10.