Loss of heterozygosity in the short arm of human chromosome 3 in sporadic lung cancer*
Lina Marcela Barrera, Biol1**, Lizeth Marelly Álvarez, Biol.1**, Miguel Ignacio Roldán, MD2, Héctor Ortega, MD3, Omar Triana, PhD4, Alonso Martínez, PhD5
* This project was funded by the Committee for the Research Development (CODI in Spanish). Initiation Acts: CPT 8840-414 and CPT 8840-415.
** When this project was begun, LMB y LMA belonged to Infection and Cancer Group.
1. Associate researcher Bacteria & Cancer Group, Department of Microbiology and Parasitology, School of Medicine, Universidad de Antioquia, Medellín, Colombia. e-mail: linabarrera07@yahoo.com lizethalvarezsalas@yahoo.com
2. Professor, Department of Pathology, School of Medicine, Universidad de Antioquia, Medellín, Colombia.
e-mail: mirope65@yahoo.com
3. Professor, Pneumology Section, Department of Internal Medicine, School of Medicine, Universidad de Antioquia, Medellín, Colombia. e-mail: hortega@une.net.co
4. Leader Chagas Group, Professor Institute of Biology, Faculty of Exact and Natural Sciences, Universidad de Antioquia, Medellín, Colombia. e-mail: otriana@gmail.com
5. Leader Bacteria & Cancer Group, Assistant Professor, Department of Microbiology and Parasitology, School of Medicine, Universidad de Antioquia, Medellín, Colombia. e-mail: amartine@une.net.co
Received for publication September 8, 2009 Accepted for publication May 7, 2010
SUMMARY
Introduction: Loss of Heterozygocity (LOH) in the short arm of human chromosome 3 (3p) is a frequent event in different types of sporadic tumors, including lung cancer (LC).
Aim: To determine 3p LOH in LC samples using 17 microsatellite markers.
Methodology: In a pilot study on volunteers, thirteen LC biopsies (tumor tissue) and 4 ml of blood (normal tissue) from the same patient were collected. DNA extraction and Polymerase Chain Reaction (PCR) were performed with 17 microsatellite markers to analyze LOH. Amplified fragments were run on 6% denaturalizing polyacrilamide gels and were visualized by using silver stain. Descriptive analysis was performed for each region on the 3p chromosome.
Results: All tumors were informative for one or more of the analyzed markers. LOH was found in one or more loci in eleven samples (84.6%). The markers with major LOH were UBE1L (23.1%), D3S1317, D3S1300, D3S1284, D3S1274, D3S3049, and D3S1577 (15.4%). Three samples showed microsatellite instability (changes in the length of the microsatellite) in different loci. The percentages of LOH for the regions of 3p were: 17.6 % for 3p24-25, 11.62% for 3p21-22, 20% for 3p13-14, and 18.42% for the 3p12 region.
Conclusions: Chromosomal regions with allelic loss were identified where probably other GSTs involved in the development of the LC are localized. It should increases sample size and marker number in order to narrow a minimal region and to identify a unknown gene involved in LC.
Keywords: Lung cancer; Microsatellite markers; Loss of heterozygosity (LOH).
Pérdida de heterocigocidad en el brazo corto del cromosoma 3 humano en cáncer esporádico de pulmón
RESUMEN
Introducción: La pérdida de heterocigocidad (LOH) en el brazo corto del cromosoma 3 (3p) humano es un evento frecuente en diferentes tipos de tumores esporádicos, incluyendo cáncer de pulmón (CP).
Objetivo: Determinar la LOH de 3p en muestras de CP, con 17 marcadores microsatelitales.
Metodología: En un estudio piloto en voluntarios, se recolectaron 13 biopsias de CP (tejido tumoral) y 4 ml de sangre periférica (tejido normal) del mismo paciente, se extrajo el ADN y se realizaron reacciones en cadena de la polimerasa (PCR) con 17 marcadores microsatelitales para analizar LOH. Los fragmentos amplificados se corrieron en geles de poliacrilamida desnaturalizante al 6% y se visualizaron por medio de la coloración de tinción de plata. El análisis descriptivo se realizó para cada región estudiada en el cromosoma 3p.
Resultados: Todos los tumores fueron informativos para uno o más de los marcadores analizados. Se encontró LOH en uno o más loci en 11 muestras (84.6%). Los marcadores con mayores LOH fueron UBE1L (23.1%), D3S1317, D3S1300, D3S1284, D3S1274, D3S3049 y D3S1577 con 15.4%. Tres muestras presentaron inestabilidad microsatelital (cambios en la longitud del microsatélite) en diferentes loci. Los porcentajes de LOH para las regiones de 3p fueron: 17.6 % para 3p24-25, 11.6% para 3p21-22, 20% para 3p13-14 y 18.4% para la región 3p12.
Conclusiones: Se identificaron regiones cromosómicas con pérdida alélica donde es probable que se localicen otros GST involucrados en el desarrollo de CP, diferentes de los ya identificados como VHL, RASSF1A, FHIT y DUTTI, entre otros. Se debe aumentar el número de muestras y de marcadores para delimitar una región mínima e identificar algún gen no descrito implicado en la carcinogénesis de pulmón.
Palabras clave: Neoplasia pulmonar (NP); Marcadores microsatelitales; Pérdida de heterocigocidad (LOH).
Lung cancer (LC) represents 12.6% (1’448,000) of new cancer cases in the world; it is the most frequent cause of mortality due to cancer in males (848,132 per year) and the second in females (330,786 per year)1. In Colombia nearly 3868 cases/year are diagnosed in a proportion of two men for one woman and there are approximately 3895 deaths per year, which places it as the third cause of deaths due to cancer2. LC originates in the respiratory epithelium and it is classified as Small-Cell Lung Cancer (SCLC) and Non-Small-Cell Lung Cancer (NSCLC). Cigarette smoking is the main risk factor causing LC and it is involved in up to 71% of deaths due to LC in Western countries3. In addition to tobacco, there are other factors involved in the development of LC related to life style and occupational risk factors that imply being in contact with carcinogens4.
Tumor Suppressor Genes (TSG) are implied in carcinogenesis, like also proto-oncogene and DNA repair genes; the existence of TSG is suspected when observing frequent loss of specific regions in a specific chromosome. If we correlate absence of locus with tumor development, it can be supposed that its presence avoids tumorigenesis. For a cell to acquire tumor phenotype, loss of TSG function is required, which may or may not be accompanied by over expression of proto-oncogene and there is also accumulation of additional mutations in other genes5,6.
Karyotypes and analysis of Loss of Heterozygosity (LOH) are routine procedures to detect DNA deletion and serve as TSG location indicators. Several authors postulating LOH occurs because there is loss of the whole chromosome due to inappropriate chromosome segregation during mitosis or that it is also produced because genetic alterations change chromosome structures. Other authors suggesting mitotic recombination and gene conversion are additional mechanisms causing LOH7. Detecting genetic changes in lung epithelium is important in order to identify patients at high risk of developing LC. Pre-neoplastic and pre-invasive lesions possess changes like LOH in 3p, 9p, 8p, and 17p arms and mutations in some oncogenes and TSG. Recent studies suggest that these changes are also observed in bronchial epithelium of normal appearance in smokers and former smokers and are even detected in sputum samples obtained from these patients, which serve as molecular biomarkers for early detection of LC8. The aim of this study is to determine LOH in human chromosome 3p in 13 samples of sporadic LC using 17 microsatellite markers.
METHODOLOGY
This pilot study was carried out with 13 volunteer patients diagnosed with sporadic LC, without chemotherapy and/or radiotherapy treatment from whom we obtained an informed signed consent previously approved by the respective ethics committees. Through a structured interview, we obtained information on their individual life styles, including cigarette and alcohol consumption. 4 ml of peripheral blood was obtained from each patient (normal control), together with a biopsy of the tumor, which was classified according to the American Joint Committee on Cancer (AJCC). 10 mg of tumor tissue were separated and homogenized with Proteinase K (Sigma, USA), and lymphocytes were isolated from peripheral blood through a centrifuge gradient of FICOLL HISTOPAQUE® 1077 (Sigma, USA). Afterwards, the DNA was extracted from the paired samples of tumor and normal tissue with the Wizard® Genomic DNA Purification kit (Promega, USA). The DNA was quantified and diluted to a 50 ng/ml concentration.
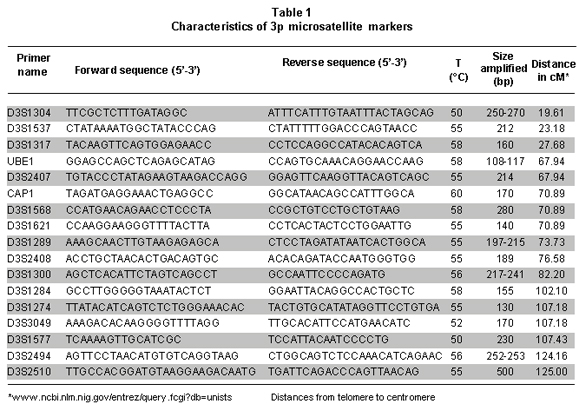
LOH in 3p was determined using 17 microsatellite markers synthesized by Integrated DNA Technologies, Inc, USA (Table 1) and optimized in previous studies9,10. Standard Polymerase Chain Reaction (PCR) was carried out in 25µl final volume containing 50 ng/ml of genomic DNA; 1X Ammonium buffer; 1.25 mM of MgCl2; 12.5 pmol of each primer pair; 0.04 mM of each deoxynucleotide (dTTP, dGTP, dCTP, and dATP); 0.5 U of Taq Recombinant polymerase (Fermentas Life Sciences, USA). Amplification was done on a PT-100 thermocycler (Sensoquest, Germany) with the following thermal profile: initial denaturation at 94°C for five min, followed by 35 cycles consisting of denaturation at 94°C for 30 s, a specific annealing temperature for each set of primer for 30 s, (Table 1), and an extension at 72°C for 1 min; followed by a final extension at 72°C for 10 min.
The amplified products were run on 6% denaturalizing polyacrilamide gels. The gels were preheated at 1700 volts for 30 min. Formamide loading buffer in a 5:1 ratio was added to the PCR products, which were denaturized at 95°C for 5 min and loading into the gel. Electrophoresis was carried out in a vertical chamber (Sequi-Gen, Bio-Rad, USA) during 4-6 hours at constant voltage depending on size of the fragment to accomplish an adequate separation. Then, silver nitrate staining was carried out. Briefly, the gel was washed in 10% acetic acid for 30 min with constant agitation, rinsed in ddH2O for 2 min; then it was incubated into 1% nitric acid during 15 min and again rinsed with ddH2O. Thereafter, the gel was submerged in a silver nitrate and formaldehyde solution for 30 min and rinsed with ddH2O for 2 min. Then, it was introduced onto a pre-cold developing solution (4ºC) of sodium carbonate, formaldehyde, and sodium thiosulphate until the bands were visualized. Finally, the reaction was stopped with 10% acetic acid and rinsed with running water. The gel was transferred to Whatman 3MM paper and vacuum dried for 90 min at 80°C. LOH was assigned if an allele was absent or exhibited an altered signal in tumor sample with relationship to normal DNA allelic ratio. A descriptive analysis was conducted for each region studied in chromosome 3p.
RESULTS
Patient characteristics are described in Table 2. Patients were in an age range between 49 and 78 years, with a mean of 67 years of age. All were active smokers from an early age, with consumption over 20 cigarettes/day (high consumption). Of these, nine individuals (69.2%) regularly consumed alcoholic beverages (moderate consumption, between 50 and 100 g/day). Two samples (15.4%) were classified as SCLC and ten (77%) were classified as NSCLC. NSCLC samples were classified according to the cellular type involved in malignant transformation; six samples were squamous cell carcinoma, two bronchogenic squamous cell carcinomas, one adenocarcinoma, and one squamous cell carcinoma. One of the samples presented non-malignant dysplasia; however, molecular study showed important changes which were considered for analyses.
All tumors were informative for one or more of the markers analyzed. In 11 of 13 samples (84.6%) LOH was found in one or more loci. The markers with major LOH were: UBE1L (23.1%), D3S1317, D3S1300, D3S1284, D3S1274, D3S3049, and D3S1577 with 15.4% (Figure 1). LOH percentages for each 3p regions were calculated as: number of LOH in the region/number of informative cases in the same region. The percentages obtained were: 17.6% for 3p24-25, 11.6% for 3p21-22, 20% for 3p13-14, and 18.4% for 3p12 region. In three samples, NT44, NT45, and NT48, microsatellite instability (MSI) and LOH were simultaneously found (Figures 2 and 3). Two squamous cell carcinoma-type NSCLC tumors did not present allelic loss for any loci. One patient, NT55, was found with allelic loss of microsatellite marker D3S1577, flanked by markers CAP1 and D3S3049, both showed allelic retention (Figure 4).
DISCUSSION
Tobacco smoking is the main risk factor for developing LC and other types of cancer11,12. Furthermore, consumption of alcoholic beverages is also an important factor in lung, mouth, larynx, pharynx, stomach, and liver tumorigenesis when consumption overpasses 100 g/day4. Patients studied were active smokers for several years (high consumption) and were moderate alcohol drinkers. A meta-analysis performed by Lubin et al.13, calculated that 25-68% of cancers associated to upper aero-digestive tract are related to alcohol consumption and the incidence increases in up to 80% when is combined with cigarette smoking. The same study reports that the combined effect of alcohol and cigarette smoking increases the risk of developing LC up to 80 times, which indicates the multiplicative effect of both. It is suggesting the first carcinogenic event is caused by tobacco and gives the affected cell a selective advantage that together with alcohol consumption favors accumulation of new cell mutations and alterations that in the end would manifest as a tumor phenotype.
In the 3p24-25 region, three LOH (18.8%) were found of the total found in 13 patients. Previous reports located and identified some genes related to tumor development as VHL, VHI, and RARb, which are flanked by microsatellites D3S1537 and D3S1317. Miyakis et al.14, demonstrated that mutations and LOH in VHL gene are not common events in NSCLC; nevertheless, they found LOH in regions adjacent to this gene in squamous cell carcinoma and adenocarcinomas at 74.6% and 49.3%, respectively. It is postulated that other still not identified genes located in this region would be genetically altered and involved in tumorigenesis. Our results are agree with the high frequency of LOH in this region; although, it is not comparable with previous study probably due to the low number of microsatellites employed to cover this 4.5 cM region.
For the 3p21-22 region, we used six microsatellite markers covering a distance of 16.58 cM, where five LOH (11.7%) were found. This region contains the Lung Cancer Tumor Suppressor Gene Region 1 (LCTSGR1), where genes as RASSF1A, HYAL2/LUCA, FUS1/TUSC2, 101F6/CYB561D2, and CPR2L/CPRL2/TUSC4, are located. Previous studies reporting RASSF1A gene presents LOH in up to 75% of NSCLC15. Other authors reported that HYAL2/LUCA, FUS1/TUSC2, 101F6/CYB561D2, and CPR2L/CPRL2/TUSC4 genes presented genetic changes (LOH, point mutations, deletions, and epigenetic changes in different types of cancers and tumor-cell lines); however, they did not identify LOH in LC biopsies5,16. The results herein reported show a high frequency of LOH; even though, we cannot guarantee that it corresponds to RASSF1A gene or to another gene located in this region, which will be the next step to study in our samples.
In 3p13-14 region with a distance of 19.9 cM between markers D3S1300 and D3S1284, four cases of LOH (20%) were identified. The most fragile site of the genome is located in this region and the FHIT gene is located here, which is involved in development of different types of cancer. Zabarosky et al.17, refer to some studies demonstrated that up to 50% of LC cases present LOH in FHIT gene. They also reported that another gene located in this region, FOXP1, presents loss of expression in 51% of cancers analyzed; however, they did not report LOH in LC samples. Our results are similar to those reported for this region. 3p12 region, contains the Lung Cancer Tumor Suppressor Gene Region 2 (LCTSGR2), where the DUTT1 gene is located; to analyze this region, we used five markers covering a 17.82-cM distance and found the greatest number of LOH, 7 (18.5%) for this study. Dallol et al.18, demonstrated that hypermethylation occurs in DUTT1 gene in up to 4% of LCs and observed LOH for some markers in 3p12 region. Our results show a smaller difference than that reported in similar studies19. For this region, patients NT43, NT51, and NT55 showed LOH in a microsatellite marker flanked by two markers that showed retention. It suggesting this region is very important in the development of tumorigenesis and there could be a yet unidentified candidate TSG (Figure 2).
There were three cases of microsatellite instability (MSI) in markers D3S2407, D3S1289, and D3S1274 (Figure 2). MSI are changes in the length of the microsatellite marker by insertions or deletions of repetitions in tumor tissue when compared with normal tissue; these alterations may occur due to defects in the DNA repair system (Mismatch Repair - MMR). Nevertheless, others studies identified microsatellite alterations like LOH and MSI in 80.5% of LC samples for chromosomes 3p, 5q, 8p, 9p, 9q, and 17q. Additionally, studies reporting genes involved in DNA repair do not present changes in their expression pattern in SCLC cellular lines and that MSI is a bio-marker for early detection of LC, mainly in NSCLC20.
In conclusion, chromosomal regions were found with high frequency of LOH where TSG involved in LC development are probably located. This work will permit conducting another study in which the number of samples is increased and comparing to a control group with similar characteristics as the cases, together with increasing the number of informative microsatellite markers for the Colombian population to narrow a minimal region to a few hundred base pairs to permit identifying some yet unknown gene important in lung carcinogenesis, because it was determined that LOH phenomenon is a critical and early genetic event for onset and development of the disease. Detecting LOH in tissues of normal appearance surrounding tumor tissue or in tissues with early stages of the disease will lead to early diagnosis and to efficient preventive and curative treatment. Finally, it must be stressed that this is a pioneering study on LOH conducted in a Colombian population.
Conflict of interest. None of the authors has conflicts of interest related to this study.
REFERENCES
1. World Health Organization. The global burden of disease: 2004 update. Geneva: WHO Press; 2008.
2. Piñeros M, Ferlay J, Murillo R. Cancer incidence estimates at the national and district levels in Colombia. Salud Publica Mex. 2006; 48: 455-65.
3. World Health Organization. Global health risks. Mortality and burden of disease attributable to selected major risks. Geneva: WHO Press; 2009.
4. Poschl G. Seitz H. Alcohol and cancer. Alcohol Alcohol. 2004; 39: 155-65.
5. Hesson LB, Cooper WN, Latif F. Evaluation of the 3p21. 3 tumour-suppressor gene cluster. Oncogene. 2007; 26: 7283-301.
6. Kuroki T, Tajima Y, Furui J, Kanematsu T. Common fragile genes and digestive tract cancers. Surg Today. 2006; 36: 1-5.
7. Ogiwara H, Kohno T, Nakanishi H, Nagayama K, Sato M, Yokota J. Unbalanced translocation, a major chromosome alteration causing loss of heterozygosity in human lung cancer. Oncogene. 2008; 27: 4788-97.
8. Breuer RH, Postmus PE, Smit EF. Molecular pathology of non-small-cell lung cancer. Respiration. 2005; 72: 313-30.
9. Martinez A, Walker RA, Shaw JA, Dearing SJ, Maher ER, Latif F. Chromosome 3p allele loss in early invasive breast cancer: detailed mapping and association with clinicopathological features. Mol Pathol. 2001; 54: 300-6.
10. Martínez A. Mapping, identification and characterization of candidate tumour suppressor gene(s) on the short arm of human chromosome 3 involved in several cancers. [PhD Thesis]. Birmingham: University of Birmingham Press, University of Birmingham; 2001.
11. Tse LA, Mang OW, Yu IT, Wu F, Au JS, Law SC. Cigarette smoking and changing trends of lung cancer incidence by histological subtype among Chinese male population. Lung Cancer. 2009. 66: 22-7.
12. Sasco AJ. Cancer and globalization. Biomed Pharmacother. 2008; 62: 110-21.
13. Lubin JH, Purdue M, Kelsey K, Zhang ZF, Winn D, Wei Q et al. Total exposure and exposure rate effects for alcohol and smoking and risk of head and neck cancer: a pooled analysis of case-control studies. Am J Epidemiol. 2009; 170: 937-47.
14. Miyakis S, Liloglou T, Kearney S, Xinarianos G, Spandidos DA, Field JK. Absence of mutations in the VHL gene but frequent loss of heterozygosity at 3p25-26 in non-small cell lung carcinomas. Lung Cancer. 2003; 39: 273-77.
15. Lerman MI, Minna JD for The international lung cancer chromosome 3p21.3 tumour suppressor genes consortium. The 630-kb lung cancer homozygous deletion region on human chromosome 3p21. 3: Identification and evaluation of the resident candidate tumor suppressor genes1. Cancer Res. 2000; 60: 6116-33.
16. Sharp TV, Al-Attar A, Foxler DE, Ding L, de A Vallim TQ, Zhang Y et al. The chromosome 3p21. 3-encoded gene, LIMD1, is a critical tumor suppressor involved in human lung cancer development. Proc Natl Aca Sci. 2008; 105: 19932-37.
17. Zabarovsky ER, Lerman MI, Minna JD. Tumor suppressor genes on chromosome 3p involved in the pathogenesis of lung and other cancers. Oncogene. 2002; 21: 6915-35.
18. Dallol A, Forgacs E, Martínez A, Sekido Y, Walker R, Kishida T et al. Tumour specific promoter region methylation of the human homologue of the Drosophila Roundabout gene DUTT1 (ROBO1) in human cancers. Oncogene. 2002; 21: 3020-28.
19. Agathanggelou A, Honorio S, Macartney DP, Martínez A, Dallol A, Rader J et al. Methylation associated inactivation of RASSF1A from region 3p21. 3 in lung, breast and ovarian tumours. Oncogene. 2001; 20: 1509-18.
20. Hansen LT, Thykjaer T, Ørntoft TF, Rasmussen LJ, Keller P, Spang-Thomsen M et al. The role of mismatch repair in small-cell lung cancer cells. Eur J Cancer. 2003; 39: 1456-67.