Detection of gene amplification in MYCN, C-MYC, MYCL1, ERBB2, EGFR, AKT2, and human papilloma virus in samples from cervical smear normal cytology, intraepithelial cervical neoplasia (CIN I, II, III), and cervical cancer*
Dabeiba Adriana García, MSc1, Ignacio Briceño, PhD2, Marcos Castillo, MD2, Fabio A. Aristizábal, PhD3
* Project financed by Universidad de La Sabana, Bogotá, Colombia.
1. Odontology Research Center, Pontificia Universidad Javeriana, Bogotá, Colombia. e-mail: garciad@javeriana.edu.co
2. Faculty of Medicine, Universidad de La Sabana, Bogotá, Colombia. e-mail: ibriceno@unisabana.edu.co mcastillo@unisabana.edu.co
3. Department of Pharmacy, Universidad Nacional de Colombia, Bogotá, Colombia. e-mail: faaristizabalg@unal.edu.co
Received for publication April 12, 2010 Accepted for publication August 20, 2010
SUMMARY
Introduction: Cervical cancer is the second most common cancer among women worldwide and the second cause of cancer mortality in women. It has been demonstrated that the process of cervical carcinogenesis displays genetic and environmental epigenetic components. Currently, research is focused on new prognosis markers like oncogene amplification.
Objectives: To perform detection of MYCN, C-MYC, MYCL1, ERBB2, EGFR, and AKT2 amplification. Additionally, to detect human papillomavirus in samples from normal cytology smear, cervical intraepithelial neoplasia (CIN) I, II, and III and cervical cancer patients.
Methods: Papillomavirus (HPV) genotyping by reverse line blot (RLB) performed and gene amplification by detection with real-time PCR with Taqman probes.
Results: HPV was present in 4% of the patients with normal cytology, 48% in CIN I, 63.6% in CIN II, 64% in CIN III, and 70.8% in cervical cancer. Genes amplified in cervical cancer were MYCN (39.1%), ERBB2 (34.7%), and MYCL1 (30.4%); showed higher amplification in high-grade lesions and cervical cancer in relation to low-grade lesions and normal cytology with statistically significant differences. Besides the genes, C-MYC, EGFR, and AKT2 were amplified in samples from patients with cervical cancer by 12%, 18%, and 13%, respectively; we did not find statistical differences.
Conclusion: Higher prevalence of gene amplification and HPV was found in high-grade cervical lesions and cervical cancer.
Keywords: HPV; Amplification; MYCN; C-MYC; MYCL1; ERBB2; EGFR; AKT2.
Colomb Med. 2011; 42: 144-53
Detección de amplificación en los genes MYCN, C-MYC, MYCL1, ERBB2, EGFR y AKT2 y del virus de papiloma humano en muestras de cepillado cervical en mujeres con citología normal y con neoplasia intraepitelial cervical (NIC) I, II, III y cáncer de cuello uterino
RESUMEN
Introducción: El cáncer cervical es el segundo cáncer más importante en mujeres a nivel mundial y es la segunda causa de muerte por cáncer en mujeres. Se ha demostrado que el proceso de carcinogénesis cervical presenta componentes tanto genéticos como epigenéticos y medio ambientales. En la actualidad, hay gran interés en la búsqueda de marcadores moleculares asociados con la progresión de esta enfermedad, uno de los posibles mecanismos y que además está poco estudiado en cáncer cervical es la amplificación génica de algunos oncogenes como la familia MYC, EGFR y AKT entre otros.
Objetivos: Detectar la amplificación génica de MYCN, C-MYC, MYCL1, ERBB2, EGFR y AKT2 además de la presencia del virus de papiloma humano en cepillados cervicales en mujeres con citología normal o con neoplasia intraepitelial cervical (NIC) I, II y III o con cáncer cervical.
Métodos: Se genotipificó mediante reverse line blot (RLB) el virus de papiloma humano (VPH) y se determinó el estado de amplificación génica de los genes mencionados mediante PCR en tiempo real utilizando sondas taqman.
Resultados: El VPH se encontró presente en 4% de las pacientes con citología normal, en 48% en NIC I, 63.6% en NIC II, 64% en NIC III y 70.8% en cáncer cervical. Los genes MYCN, MYCL1 y ERBB2 mostraron mayor amplificación en lesiones de alto grado y cáncer con diferencias estadísticamente significativas a las lesiones de bajo grado y citología normal, en 39.1%, 34.7% y 30.4% respectivamente. Además, se encontraron amplificados los genes C-MYC, EGFR y AKT2, en muestras de pacientes con cáncer cervical, en 12%, 18% y 13% respectivamente. Sin embargo, no se observaron diferencias estadísticamente significativas con respecto a las lesiones de alto y bajo grado y citología normal.
Conclusión: En las lesiones de alto grado como en cáncer cervical, se encuentra mayor prevalencia del virus al igual que se detectan mayor cantidad de alteraciones genéticas como la amplificación génica.
Palabras clave: VPH; Amplificación génica; MYCN; C-MYC; MYCL1; ERBB2; EGFR; AKT2.
Colomb Med. 2011; 42: 144-53
Cervical cancer is one of the most important diseases worldwide; in Colombia, the estimated incidence is 21.5 cases per 100,000 inhabitants1. The natural biology of the cervical cancer process begins with noninvasive squamous lesions that progress to cervical intraepithelial neoplasia (CIN) I, II, and III. In some cases, the lesions progress to invasive carcinoma2. However, it is not yet known what lesions progress and which of them revert their state, nor the mechanisms of tumor progression in this type of neoplasia. No one has found a molecular marker that is significantly associated with the progression or clinical outcome of the disease.
Among prognostic markers in cervical cancer amplification, hTERT has been proposed. The gene amplification activates the oncogenic potential of cellular protooncogenes, which is a reflection of genetic instability of tumor cells and is a particular feature of an aggressive series of tumors3. The term Gene amplification refers to the increased number of gene copies as a local effect. Amplification of specific genes has been implicated in an increasing variety of cell adaptive responses to environmental stress and it is one of the mechanisms by which the cell responds to the demand for the synthesis of specific gene in amounts exceeding the transcriptional capacity of a single copy of a gene4. Amplification is an non-programmed aberration only seen in response to the exposure of certain cytotoxic drugs and as an associated phenomenon with tumorigenesis.
Among the techniques used to detect gene amplification are fluorescent in situ hybridization (FISH), array-based comparative genomic hybridization (array-CGH), and real-time PCR (qPCR). Malakh et al., in 2008, showed a strong correlation among these three methods, close to 99%, to detect a number of MYCN copies of brain cancer5.
In cervical cancer, amplification of the EGFR oncogene has been observed, which is considered a marker of poor prognosis, mainly in breast and lung cancer. Although in cervical cancer only a low percentage of such is amplified, this amplification occurs mainly in adenocarcinomas also correlating with poor prognosis6. Other genes such as C-MYC and MYCN have been found amplified up to five times more in 40% of cervical cancer, also suggesting a poor prognosis7. However, there are few studies evaluating gene amplification in cervical cancer of genes MYCL1, ERBB2, and AKT2. For this reason, in this study we sought to evaluate gene amplification of EGFR, C-MYC, MYCN, MYCL1, ERBB2, and AKT2 in both women with normal cytology and patients with preneoplastic and neoplastic lesions of cervical cancer.
METHODS
Population and samples. This study is descriptive, cross-sectional and observational; the number of patients was defined by convenience sampling and was collected during an eight-month period in a hospital in the city of Bogotá. Cervical brush samples were taken by trained personnel, the patients included in the study were 50 with negative cytological results and 96 patients with pathologically confirmed diagnosis (25 with CIN I, 22 CIN II, 25 CIN III, and 24 with cervical cancer). These patients had not started any treatment and agreed to participate voluntarily in the study by signing an informed consent.
The cervical smears were taken in 1X PBS and 0.05% thimerosal. The sample was divided into two aliquots, one of them was centrifuged at 3000 g for 10 minutes, 1 ml of 10 mM tris-HCl buffer (pH 8.3) was added to the obtained precipitate; after that, the sample was heated for 10 minutes and centrifuged at 3000 g for one minute, the supernatant was used directly to perform PCR of β-globin and HPV genotyping by Reverse Line Blot (RLB). In the other aliquot, DNA was isolated by phenol-chloroform method for analysis of gene amplification via real-time PCR.
DNA quality. The supernatants of cervical smears were used as templates to amplify the β-globin gene by using PCO3 (sense) 5 'ACACAACTGTGTTCAC TAGC 3' and PCO4 (antisense) 5 'CAACTTCATC CACCTTCACC 3' primers. The final concentrations used were 10 mM Tris-HCl, 50 mM KCl, 0.1% triton X-100, 3 mM MgCl2, 7.5 pmol of each primer, 1 U Taq polymerase and 200 um of each dNTPs. Amplification was performed in the thermocycler MyCyclerTM (BioRad Laboratories, USA), conditions were 1 cycle at 95°C for 3 minutes and 40 cycles at 95°C for 20 seconds, 58°C for 20 seconds, and 72°C for 20 seconds. PCR products were analyzed by electrophoresis on 2% agarose and stained with ethidium bromide 10 mg/ml, the expected product was 110 bp. The positive control was lymphocyte DNA with approximately 25 ng, and PCR mixture as negative control. To find the limit of detection, dilutions were made in base 2 of lymphocyte DNA of 25 ng, and found screening to the sixth dilution equivalent to 390 pg.
HPV genotyping. We used the method described by Van Der Brule in 20028, which is based on the reverse hybridization assay, using a miniblotter system on the PCR product using primers GP5+(S) 5'TTTGTTACT GTGGTAGATACTA3' and GP6+(A) 5'GAAAAAT AAACTGTAAATCATA3'+Biotin. This genotyping method is called Reverse Line Blot (RLB), which allows identifying 37 HPV genotypes. The specific viral types were: HPV 6, 11, 16, 18, 26, 31, 33, 34, 35, 39, 40, 42, 43, 44, 45, 51, 52, 53, 54, 55, 56, 57, 58, 59, 61, 66, 68, 70, 72, 73, 82, 83, 84, 82, 71, 81, and CP6108. Plasmids of HPV 16 and HPV 18, donated by the DKFZ (German Cancer Research Center, Heidelberg, Germany), were used as positive controls; the detection limit for these two viral types was 1 fg, and as a negative control we used the PCR mixture.
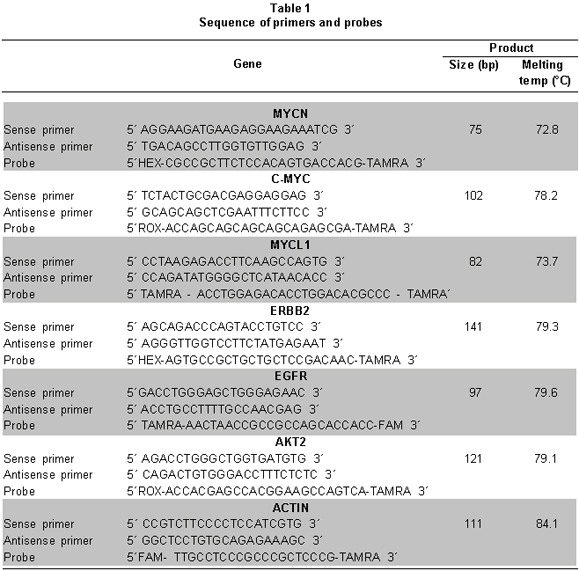
Analysis of MYCN, C-MYC, MYCL1, ERBB2, EGFR, AND AKT2 gene amplification. We designed the primers and TaqMan probes to amplify each of the genes (MYCN, C-MYC, MYCL1, ERBB2, EGFR, and AKT2) and as a reference the Actin (ACTB) gene was selected by using the Primer ExpressTM software - Applied Biosystems, Foster City, CA. Table 1 summarizes the sequences of each of the primers and probes used.
A Taqman® Universal PCR Master Mix (Applied Biosystems, Foster City, CA) PCR kit was used for real-time PCR. The final concentrations used were: 1X Master mix, each primer 50 nm, 50-nm probe, and a sample using DNA extracted with phenol-chloroform-isoamyl; each sample was evaluated in triplicate. In the negative control, water was added to the PCR mixture instead of the sample.
PCR conditions were 1 cycle at 95°C for 10 minutes followed by 40 cycles at 95°C for 15 seconds, 61°C for 1 minute, and 72°C for 20 seconds (Applied Biosystems 7500 Real-Time PCR System). We performed a standard curve for the actin or reference gene (ACTB) using base 10 dilutions of DNA from lymphocytes (15 ng to 15 fg); the limit of detection of this gene was 150 fg. The calculation to determine gene amplification was 2 (actin CT-CT target gene).
Ethical considerations. The study respected the ethical and legal aspects of research on human beings, as stipulated in resolution No. 008430 of 1993 from the Ministry of Health of the Republic of Colombia and according to the Helsinki declaration of 1975 as amended in 2004.
Patients included in this study voluntarily agreed to participate in the study after reading and understanding the informed consent, which was approved by the ethics committee at Colombia's Universidad Nacional. In addition, each patient enrolled in the study completed, with the help of the doctor taking the sample, a survey that included demographic characteristics and risk factors associated with the development of cervical cancer.
Statistical analysis. A database was developed with information from every patient, in .xls format (Microsoft Excel®. Information processing was performed by using a univariate descriptive analysis, making tables and plots for qualitative variables, such as the presence or absence of HPV and viral type. The multiple comparisons among types of injuries and analyzed gene amplification were performed by using Bonferroni's multiple comparison and variance analysis for (one-way) using the SPSS® software.
RESULTS
The socio-demographic characteristics and risk factors associated with the development of cervical cancer in the population studied are summarized in Table 2; it was observed that 53.4% of the population was younger than 34 years of age, and from this percentage up to 70.5% belonged to groups of women with normal cytology and patients with CIN I, while only 10.95% of the population studied were older than 55 years of age; 62.5% of this percentage belonged to patients with cervical cancer. With regard to the social stratum, 39.04% belonged to a lower stratum. A total of 46.6% of the population reportedly had their first sexual relationship when they were less than 18 years of age. With respect to the number of sexual partners, 66.4% of the respondents reported having had 1 to 2 partners. Some 82.2% of the women have been pregnant and among them 71.9% had their first childbirth when they were younger than 24 years of age. Finally, 48.6% do not use any contraception method, 44.5% had a history of cancer in a family member to the third degree of consanguinity and of these cases only 7.5% had a history of cervical cancer.
All samples showed a good quality, assessed by PCR amplification, of β-globin gene (Figure 1).
Genotyping results of the samples are summarized in Table 3. In general, HPV-positive patients with normal cytology were 4% for CIN I 48%, 63.6% CIN II, 64% CIN III and 70.8% in cervical cancer. Table 3 shows the distribution of viral types according to the type of injury. Of the HPV types detected, 83.6% were in the high-risk group and 16.4% were low-risk HPV types; viral type 16 was present in 31.6%, followed by HPV 18 with 15.2%, HPV 58 in 13.9%, HPV 31 and 52 with 5%, and HPV 56 with 3.8%. HPV types 33, 35, 42, 45, 59, 66, and 73 were frequent at 2.5% each, and HPV types 40, 51, 68, 70, and iso39 were found in 1.26% each. In 29.5% of HPV-positive patients mixed infection was present.
Regarding the MYCN gene amplification, an amplification trend was observed in samples with high-grade lesions compared to those from normal cytology, they also showed an amplification of 8.3% for CIN I, 27% for CIN II, 16% for CIN III and 39.1% for cancer samples; all percentages were calculated with respect to samples from patients with a normal cytology result (Figure 2). Significant statistical differences were observed between groups of normal cytology and CIN I with respect to the cancer group with p=0.0005.
For C-MYC, there is no difference of amplification in samples with some type of CIN or cancer compared to those from normal cytology, only a slight gain of 12% for cancer samples when compared with samples from patients with a normal cytology result (Figure 3). However, no significant statistical differences were noted among groups with p=0.3572.
With respect to the amplification of MYCL1, an amplification trend was observed in samples with high-grade lesions-mainly CIN II and cancer compared to those from normal cytology and CIN I; they also showed an amplification of 8.3% for CIN I, CIN II 18%, 16% for CIN III and 34.7% for cancer samples, all percentages were calculated with respect to samples from patients with a normal cytology result (Figure 4). Significant statistical differences were observed among groups with low-grade lesions compared to those with high-grade lesions and the cancer group with p=0.0046.
With ERBB2, amplification was observed mainly in cancer samples compared to those from normal cytology and CIN I and CIN II, besides amplification was observed only in CIN III in 4% and 30.4% of cancer, all percentages were calculated with respect to samples from patients with a normal cytology result (Figure 5). Significant statistical differences were observed among groups with normal cytology, CIN I, CIN III, and CIN III with respect to the cancer group with p=0.001.
In the EGFR gene, a slight tendency for amplification was observed in samples with high-grade lesions and cancer compared to those from normal cytology, they also showed amplification of 9% for CIN I, 14% for CIN II, 13% for CIN III, and 18% for cancer samples; all percentages were calculated with respect to samples from patients with normal cytology results (Figure 6). There were no statistically significant differences among groups with p=0.071.
In AKT2, although there were no statistically significant differences (p=0.6166) among groups, there was a tendency for amplification in high-grade lesions (CIN II and CIN III) and cervical cancer, with respect to low-grade lesions (CIN I ) and normal cytology (Figure 7). You can see a 4% gain for CIN I, 5% for CIN II, 8% for CIN III, and 13% for cancer samples; all percentages were calculated with respect to samples from patients with a normal cytology result.
Finally, when comparing samples with gene amplification and the presence of high-risk HPV types, we found that over 62% of the samples with MYCN, AKT2, C-MYC or ERBB2 amplification also had high-risk HPV types. In more than 50% when the genes, MYCL1 and EGFR were amplified; they also had high-risk HPV types (Table 4).
In literature, there is great variability of epidemiological data on the presence of viral DNA in patients with normal cytology, prevalence ranging from 5.2% in Spain to 33.3% found in Africa2. The frequency of HPV found in such patients in this study was 4%, similar to the 4.6% found by Jacobs et al.9 For patients with histopathological diagnosis of CIN I, the frequency we found was 48%, close to what Molano et al.10, found in 2002, which was 55.7%. For patients with high-grade lesions: CIN II and CIN III, the present study found VPH frequencies of 63.6% and 64%, respectively, similar to what Schiffman and collaborators described, who found them in 76%10. Finally, in cervical cancer patients we found a rate of HPV infection of 70.8%, correlating with what was described by Muñoz et al.2 for Colombia at 74.4%. In the present study, we found mixed infection in 29.5% of HPV-positive patients, this correlates with data from Cobo et al.11 where they observed the presence of mixed infections by 19%.
Gene amplification is an unscheduled aberration that has been seen only in response to the exposure to certain cytotoxic drugs and an associated phenomenon with tumorigenesis4. According to the Cancer Genome Project at the Sanger Institute12, the genes most commonly found to be amplified and associated with cancer are: AKT2, ALK, EGFR, ERBB2, MDM2, Mdm4, MITF, MYC, MYCL1, MYCN, and REL. In cervical cancer, a few studies assess gene amplification in different stages of cervical carcinogenesis, in this case CIN I, CIN II, and CIN III. In relation to cervical cancer, gene amplification of EGFR, MYC, and MYCN has been observed.
Analysis of gene amplification can be performed by FISH, array-CGH, and qPCR; the last used in this study offers more advantages given its accuracy, lower cost, speed, ease of interpretation, and because it requires less DNA. In addition, these three techniques have a correlation close to 99%5.
Regarding the MYCN gene in samples from patients with cervical cancer, the present study found an amplification of 39.1% compared to normal cytology results, similar to the results by Kinoshita et al.7, who described MYCN amplified through slot-blot, they observed amplification up to five times in 40% of 15 cervical cancer samples tested in Japan, suggesting poor prognosis. However, in another study by Mitra et al.13, where they evaluated 50 samples of cervical cancer in India by southern-blot, they did not find amplification of this gene.
In relation to the status of C-MYC, this was found amplified in 12% of the samples from cervical cancer patients, compared to samples from patients with normal cytology. In samples with lesions of CIN I, CIN II, and CIN III this gene was not amplified. Nevertheless, there is a wide divergence of results; for example, Baker et al.14, found it amplified in 31% of 44 cervical cancer samples from US patients. Golijow et al.15, working with 105 patients from Argentina with low- and high-grade intraepithelial lesions, through qPCR found amplification of this gene in 17% and 41%, respectively. On the contrary, Mitra et al.13, found no C-MYC amplification in cervical cancer samples.
MYCL1 amplification in this study was present in 34.7% of cancer samples compared to samples from patients with normal cytology. In cervical cancer, a few studies evaluate MYCL1 amplification in preneoplastic and neoplastic lesions of the cervix, one of these studies was conducted by Mitra et al.13, finding this gene amplified in 2% of 50 samples with cervical cancer. Nontheless, in other types of tumors such as lymphomas, there was amplification of this gene in 43% (3 samples of the 7 tested) by using microarrays16.
Another important oncogene found amplified in cancer was ERBB2; the present study detected amplification in 30.4% of the samples from patients with cervical cancer, values similar to those found by Sharma et al.17 who observed amplification in 36.6% of 60 samples with cervical cancer assessed by in situ hybridization and Zhang et al.18 who identified amplification in 29% of 84 samples tested by FISH.
In this study, there was amplification of the EGFR gene in 18% of cervical cancer samples, but few studies evaluate this gene amplification in cervical cancer and premalignant lesions. Similar to our results, Kersemaekers et al.19, in the Netherlands described EGFR amplification in this type of cancer by Sothern blot in 12.5% of the 136 cases assessed.
Taking into account that AKT2 has been found amplified in several types of cancer, including head and neck in 30% of the 117 patients evaluated from Spain by qPCR20, the present study found this gene amplified in 13% of cervical cancer samples; although until now there are no studies evaluating the amplification of this gene in cervical cancer or precancerous lesions like CIN I, CIN II, and CIN III.
According to the results observed when comparing the presence of high-risk HPV viral types and the presence of gene amplification, we observed that the presence of these viral types of HPV might somehow be related to the susceptibility of gene amplification; given that it has been observed that HPV infection increases the risk of genetic instability18. The number of cases included in this study is too limited to explain the relationship of such susceptibility. Nowadays few studies simultaneously evaluate gene amplification and high-risk HPV viral types. Zhang et al.18 with 90 cervical cancer patients from Sweden, found a strong relationship between HPV 16 and / or 18 and gene amplification of TERT and C-MYC by FISH.. Therefore, we suggest evaluating these two events in a study including more cases.
In conclusion, we observed that in general cervical cancer and high-grade lesions there was greater HPV frequency and more gene amplification. In addition, MYCN, ERBB2, and MYCL1 amplification could be candidates as potential prognostic markers in cervical cancer, because in high-grade lesions and cancer it is more common to find these genes amplified with respect to low-grade lesions. However, it requires a greater number of cases to corroborate these results.
ACKNOWLEDGMENTS
We thank the faculty of medicine at Universidad de La Sabana for their financial support for the implementation of this work and the departments of pharmacy and biotechnology and the common machine laboratory at Universidad Nacional de Colombia (Bogotá, Colombia) for their facilities to conduct this study, and especially to all the patients who agreed to participate in the study.
Conflict of interest. None of the authors has conflicts of interest related to this study.
REFERENCES
1. Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010; 127: 2893-917.
2. Muñoz N, Bosch FX, Sanjose S, Herrero R, Castellsague X, Shah V, et al. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003, 348: 518-27.
3. Savelyeva L, Schwab M. Amplification of oncogenes revisited: from expression profiling to clinical application. Cancer Letters. 2001; 167: 115-23.
4. Brison O. Gene amplification and tumor progression. Biochim Biophys Acta. 1993; 1155: 25-41.
5. Malakh SG, Korshunov A, Stroganov AM, Poltaraus AB. Fast detection of MYCN copy number Alterations in brain neuronal tumors by real-time PCR. J Clin Lab Anal. 2008; 22: 123-30.
6. Mark HF, Feldman D, Das S, Sun CL, Samy M, Lathrop J. HER-2/neu oncogene amplification in cervical cancer Studied by fluorescent in situ hybridization. Genet Test. 1999; 3: 237-42.
7. Kinoshita M, Ikei N, Shin S, Inui S, Hirao T, Aono T. Relationship between human papillomavirus and oncogenes (C-MYC, N-MYC) amplification in human cervical Cancers. Nippon Gan Chiryo Gakkai Shi. 1990; 25: 2828-35.
8. van den Brule A, Schouls L. GP5 +/6 +PCR reverse line blot followed by enables rapid analysis and high-throughput identification of human papillomavirus genotype. J Clin Microbiol. 2002, 40: 779-87.
9. Jacobs MV, Walboomers JM, Snijders PJ, Voorhorst FJ, Verheijen RH, Fransen-Daalmeijer N, et al. Distribution of 37 HPV types mucosotropic in women with cytologically normal cervical smears: the age-related patterns for high-risk and low-risk types. Int J Cancer. 2000; 87: 221-7.
10. Schiffman MH, Bauer HM, Hoover RN, Glass AG, Cadell DM, Rush BB, et al. Epidemiologic evidence showing that human papillomavirus infection causes most cervical intraepithelial neoplasia. J Natl Cancer Inst. 1993; 85: 958-64.
11. Cobo F, Concha A, Ortiz M. Human papillomavirus (HPV) type distribution in females with abnormal cervical cytology. a correlation with histological study. Open Virol J. 2009; 3: 60-6.
12. Cancer Gene Census. Amplified Cancer Genes. Trust Sanger Institute. [February 2010]. Cambridge: URL: http://www.sanger.ac.uk/genetics/CGP/Census/amplification.shtml.
13. Mitra AB, Murty VVVS, Pratap M, Sodhani P, Chaganti SK. ERBB2 (HER2/neu) oncogene is frequently amplified in squamous cell carcinoma of the uterine cervix. Cancer Res 1994; 54: 637-9.
14. Baker VV, Hatch KD, Shingleton HM. Amplification of the c-myc proto-oncogene in cervical carcinoma. J Surg Oncol. 1988; 39: 225-8.
15. Golijow CD, Abba MC, Mouron SA, Gómez MA, Dulout FN. C-MYC gene amplification detected in preinvasive intraepithelial cervical lesions Int J Gynecol Cancer. 2001; 11: 462-5.
16. Mao X, Orchard G, Lillington DM, Russell-Jones R, Young BD, Whittaker SJ. Amplification and overexpression of JUNB is associated with primary cutaneous T-cell lymphoma. Blood. 2003; 101: 1513-9.
17. Sharma A, Pratap M, Sawhney VM, Khan IU, Bhambhani S, Mitra AB. Frequent amplification of C-erbB2 (HER-2/neu) oncogene in cervical carcinoma as detected by non-fluorescence in situ hybridization technique on paraffin sections. Oncology. 1999; 56: 83-7.
18. Zhang A, Maner S, Betz R, Angstrom T, Stendahl U, Bergman F, et al. Genetic alterations in cervical carcinomas: Frequent low-level amplifications of oncogenes are associated with human papillomavirus infection. Int J Cancer. 2002; 101: 427-33.
19. Kersemaekers AM, Fleuren GJ, Kenter GG, Van den Broek LJ, Uljee SM , Hermans J , et al. Oncogene alterations in carcinomas of the uterine cervix: overexpression of the epidermal growth factor receptor is associated with poor prognosis. Clin Cancer Res. 1999; 5: 577-86.
20. García JM, García D, Muñoz C, Herrero A, Rodrigo JP, Suárez C, et al. Frequent genetic and biochemical alterations of the PI 3-K/AKT/PTEN pathway in head and neck squamous cell carcinoma. Int J Cancer. 2004; 114: 242-8.